Chapter: Essential Microbiology: Microbial Genetics
What exactly do genes do?
What exactly do genes do?
At the start of the 20th century Archibald Garrod had proposed that inherited disorders such as alkaptonuria may be due to a defect in certain key metabolic enzymes, thus offering for the first time an explanation of how genetic information is expressed. His ideas were not really developed however, until the work of George Beadle and Edward Tatum in the 1940s, whose experiments with the bread mould Neurospora led to the formulation of the one gene, one enzyme hypothesis.
Although now acknowledged to be somewhat over-generalised, this model proved useful in the years when the molecular basis of gene action was being elucidated Having established that genes are made of DNA, and having a model for the structure of DNA that explained how it was able to copy itself, the way was open in the 1950s for scientists to work out the mechanism by which the information encoded in a DNA sequence was converted into a specific protein.
How does a gene direct the synthesis of a protein?
You may recall that both DNA and proteins are polymers whose ‘build-ing blocks’ (nucleotides and amino acids respectively) can be put together in an almost infinite number of sequences. The sequence of amino acids making up the primary structure of a protein is determined by the sequence of nucleotides in the particular gene responsible for its production. It does this not directly, but through an intermedi-ary molecule, now known to be a form of RNA called messenger RNA (mRNA). It is this intermediary that carries out the crucial task of passing the information encoded in the DNA sequence to the site of protein synthesis. This unidirectional flow of information can be summarised:
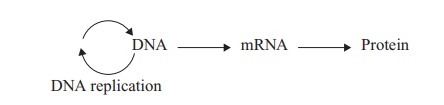
and is often referred to as the Central Dogma of biology, because of its applicability to all forms of life. Proposed by Crick in the late 1950s, this is still accepted as being a true model of the basic events in protein synthesis. Sometimes the message encoded in DNA is transcribed into either ribosomal RNA (rRNA) or transfer RNA (tRNA); these types of RNA are not translated into proteins but represent end-products in themselves.
The conversion of information encoded as DNA into the synthesis of a polypeptide chain occurs in two distinct phases (Figure 11.5); first the ‘message’ encoded in the
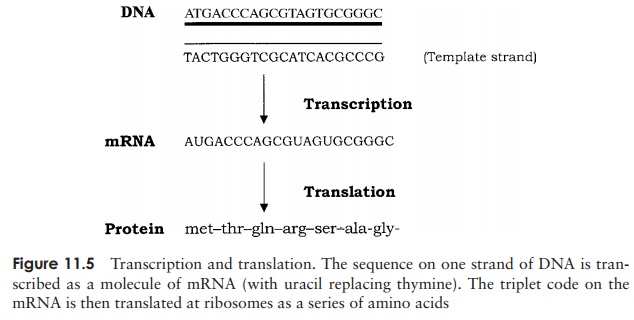
DNA sequence of a gene is converted to mRNA by tran-scription, then this directs the assembly of a specific se-quence of amino acids during translation. We shall dis-cuss how this happens shortly, but first we need to con-sider the question: how does the sequence of nucleotides in a gene serve as an instruction in the synthesis of pro-teins?
The genetic code
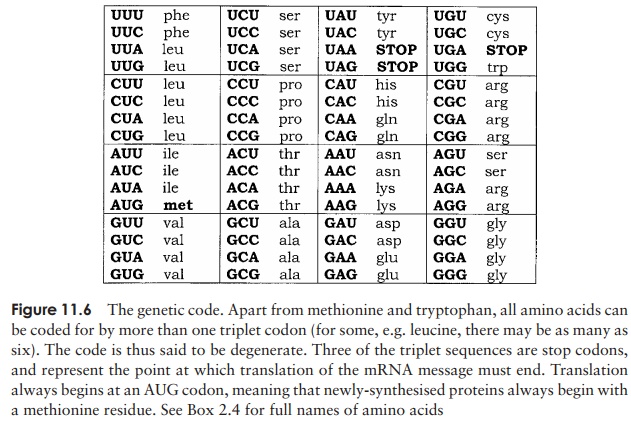
Messenger RNA carries information (copied from a DNA template) in the form of a genetic code that directs the synthesis of a particular protein. The nature of this codewas worked out in the early 1960s by Marshall Nirenberg, Har Gobind Khorana and others. The message encoded in a gene takes the form of a series of triplets (codons). Of the 64 possible three-letter combinations of A, C, G and U, 61 correspond to specific amino acids, while the remaining three act as ‘stop’ messages, indicating that reading of the message should cease at that point (Figure 11.6). It is essential that the reading of the message starts at the correct place, otherwise the reading frame(groups of three nucleotides) may become disrupted. This would lead to a completely inappropriate sequence of amino acids being produced. Frameshift mutations have this effect. Since there are only 20 amino acids to account for, it follows that the genetic code is degenerate, that is, a particular amino acid may be coded for by more than one triplet. Amino acids such as serine and leucine are encoded by as many as six alternatives each, whilst tryptophan and methionine are the only amino acids to have just a single codon (Figure 11.6).
Transcription in procaryotes
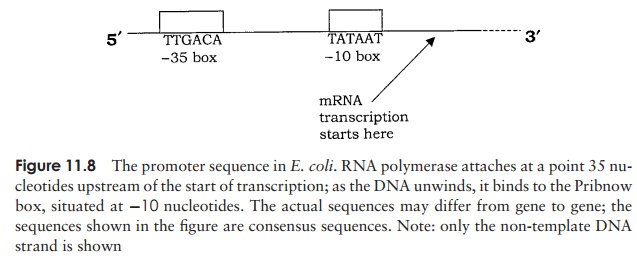
In the first phase of gene expression, one strand of DNA acts as a template for the production of a complementary strand of RNA. In the outline that follows, we shall describe how mRNA is synthesised, but remember that sometimes the product of tran-scription is rRNA or tRNA. An important point to note is that the coding strand is not the same for all genes; some are encoded on one strand, some on the other. Whereas in DNA replication the whole molecule is copied, an RNA transcript is made only of specific sections of DNA, typically single genes. The enzyme RNA polymerase, unlike DNA polymerase, is able to use completely single-stranded material, that is, no primer is required. It is able to synthesise an mRNA chain from scratch, according to the coded sequence on the template, and working in the 5 to 3 direction (Figure 11.7). In order to do this the RNA polymerase needs instructions for when to start and finish. First, it recognises a short sequence of DNA called a promoter, which occurs upstream of a gene. A protein cofactor called sigma (σ ) assists in attachment to this, and is released
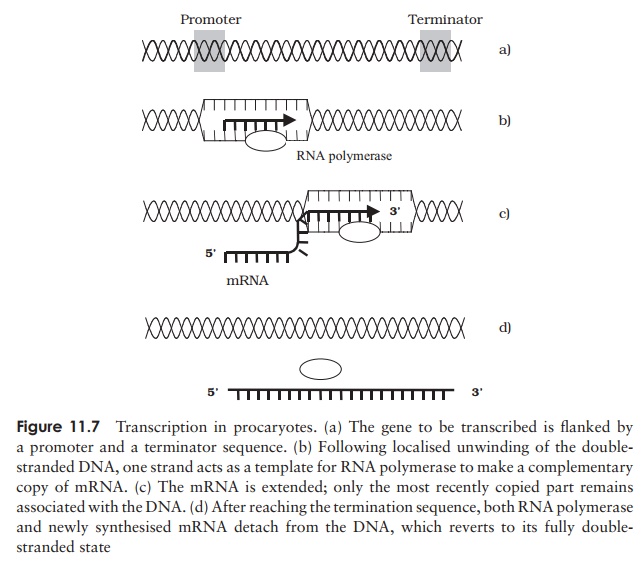
again shortly after transcription commences. The promoter tells the RNA polymerase where transcription should start, and also on which strand. The efficiency with which a promoter binds the RNA polymerase determines how frequently a particular gene will be transcribed. The promoter comprises two parts, one 10 bases upstream (known as the Pribnow box), and the other 35 bases upstream (Figure 11.8). RNA polymerase binds to the promoter, and the double helix of the DNA is caused to unwind a little at a time, exposing the coding sequence on one strand. Ribonucleotides are added one by one to form a growing RNA chain, according to the sequence on the template; this occurs at a rate of some 30–50 nucleotides per second. Remember that RNA has uracil rather than thymine, so that a ‘U’ is incorporated into the mRNA when-ever an ‘A’ appears on the template. Transcription stops when a terminator sequence is recognised by the RNA polymerase; both the enzyme and the newly synthesised mRNA are released. Unlike the promoter sequence, the terminator is not transcribed. Some termination sequences are dependent on the presence of a protein called the rho factor (ρ). Groups of bacterial proteins having related functions may have their genes grouped together. Only the last one has a termination sequence, so a single, contiguous mRNA is produced, encoding several proteins (polycistronic mRNA).
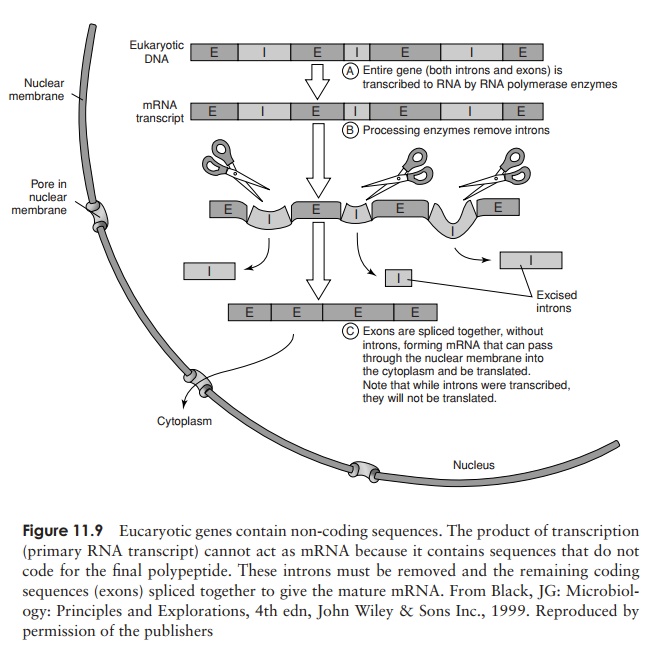
Transcription in eucaryotes proceeds along similar lines, but with certain differences. The most important of these is that in eucaryotes, the product of transcription does not act directly as mRNA, but must be modified before it can undergo translation. This is because of the presence within eucaryotic genes of DNA sequences not involved in coding for amino acids. These are called in-trons (c.f. coding sequences = exons), and are removed to give the final mRNA by aprocess of RNA splicing (Figure 11.9).
Translation
The message encoded in mRNA is translated into a sequence of amino acids at the ribosome. The ribosomes are not protein-specific; they can translate any mRNA to synthesise its protein. Amino acids are brought to the ribosome by a transfer RNA (tRNA) molecule. Each tRNA acts as an adaptor, bearing at one end the complementary sequence for a particular triplet codon, and at the other the corresponding amino acid (Figure 11.10). It recognises a specific codon and binds to it by complementary base pairing, thus ensuring that the appropriate amino acid is added to the growing peptide chain at that point. Enzymes called aminoacyl-tRNA synthetases ensure that each tRNA is coupled with the correct amino acid in an ATP-dependent process.
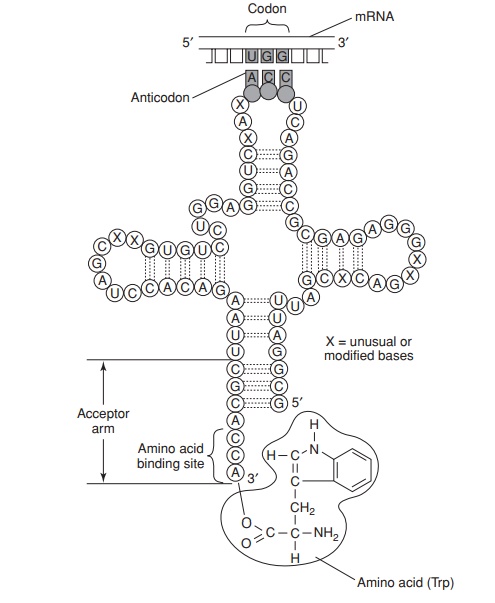
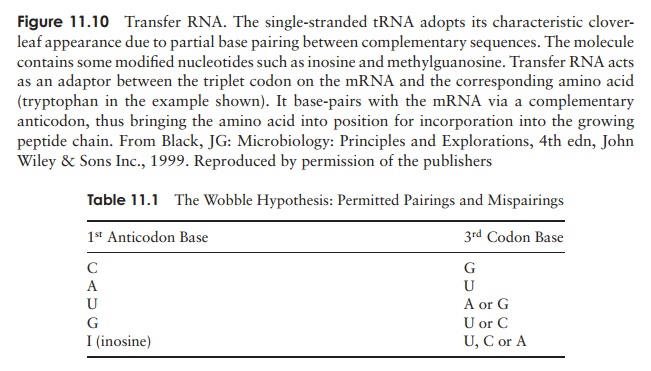
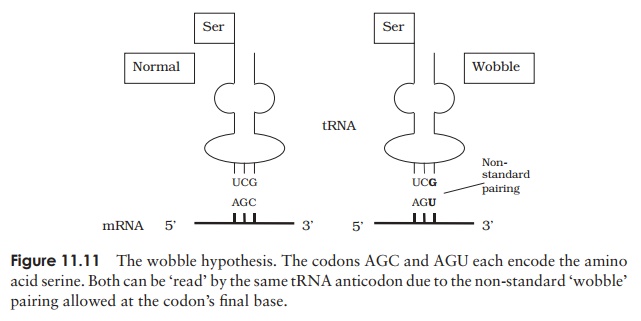
There is at least one type of tRNA for each amino acid, each with a three base anticodon, enabling it to bind to the complementary triplet sequence on the mRNA.However, there is not a different anticodon for each of the 61 possible codons, in fact there are less than 40. To explain this, the wobble hypothesis proposed that certain non-standard pairings are allowed between the third nucleotide of the codon and the first of the anticodon (Table 11.1). This means that a single anticodon may pair with more than one codon (Figure 11.11).
Translation starts when the small ribosomal subunit binds to a specific sequence on the mRNA upstream of where translation is to begin. This is the ribosome bind-ing site; in procaryotes this sequence is AGGAGG (the Shine–Dalgarno sequence).
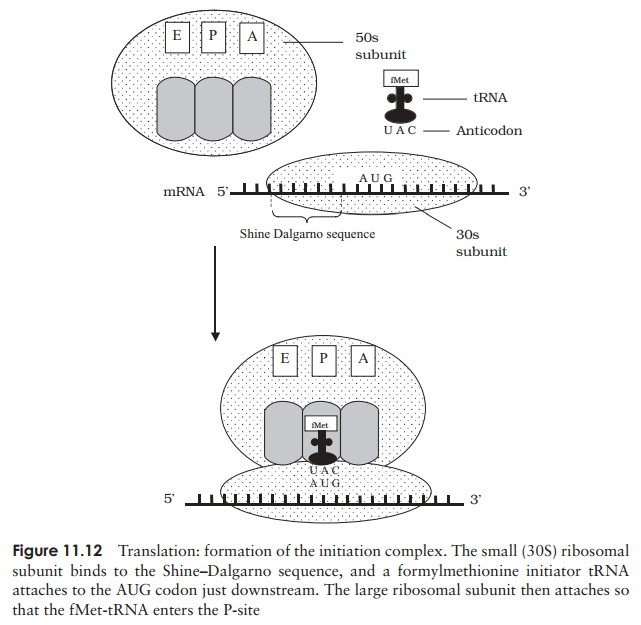
This sets the ribosome in the correct reading frame to read the message encoded on the mRNA. A tRNA car-rying a formylmethionine then binds to the AUG start codon on the mRNA. The large ribosomal subunit joins, and the initiation complex is complete (Figure 11.12). Proteins called initiation factors help to assemble the ini-tiation complex, with energy provided by GTP.
The positioning of the large subunit means that the initiation codon (AUG) fits into the P-site, and the next triplet on the mRNA is aligned with the A-site. Elon-gation of the peptide chain (Figure 11.13) starts whena second tRNA carrying an amino acid is added at theA-site.Peptidyl transferase activity breaks the link between the first amino acid and its tRNA, and forms a peptide bond with the second amino acid. The catalytic action is due partly to the ribozyme activity of the large subunit rRNA. The ribosome moves along
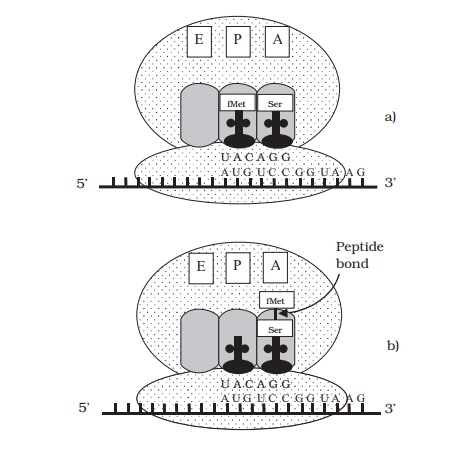
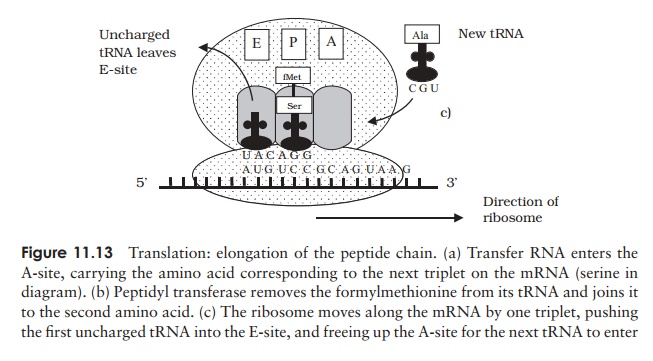
by one triplet so that the second tRNA occupies the P-site. The first tRNA is released from its amino acid, and passes to the E-site before being released from the ribosome. A third aminoacyl tRNA moves into the A-site, corresponding to the next codon on the mRNA. Elongation continues in this way until a stop codon is encountered (UAG, UAA, UGA). Release factors cleave the polypeptide chain from the final tRNA and the ribosome dissociates into its subunits.
Related Topics