Chapter: Essential Microbiology: Microbial Genetics
DNA replication in procaryotes
DNA replication in procaryotes
You may recall that bacteria multiply by a process of binary fission; before this occurs, each cell must duplicate its genetic information so that each daughter cell has a copy.
DNA replication involves the action of a number of specialised enzymes:
· Helicases
· DNA topoisomerases
· DNA polymerase I
· DNA polymerase III
· DNA primase
· DNA ligase.
Replication begins at a specific sequence called an ori-gin of replication. The two strands of DNA are caused toseparate by helicases (Figure 11.2), while single-strandedDNA binding proteins (SSB) prevents them rejoining.Opening out part of the double helix causes increased tension (supercoiling) elsewhere in the molecule, which is relieved by the enzyme DNA topoiosomerase (some-times known as DNA gyrase). As the ‘zipper’ moves along, and more single stranded DNA is exposed, DNApolymerase III adds new nucleotides to form a comple-mentary second strand, according to the rules of base-pairing. DNA polymerases are not capable of initiating the synthesis of an entirely new strand, but can only extend an existing one. This is because they require a free 3 -OH group onto which to attach new nucleotides. Thus, DNA polymerase III can only work in the 5 –3direction. A form of RNA polymerase called DNA pri-mase synthesises a short single strand of RNA, whichcan be used as a primer by the DNA polymerase III. (Figure 11.2).
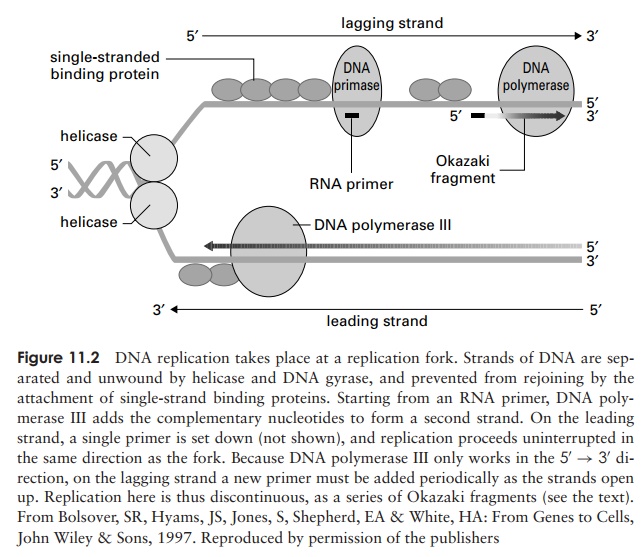
When replication occurs, complementary nucleotides are added to one of the strands (the leading strand) in a continuous fashion (Figure 11.2). The other strand (the lagging strand), however, runs in the opposite polarity; so how is a complementary sequence synthesised here? The answer is that DNA polymerase III allows a little unwinding to take place and then, starting at the fork, works back over from a new primer, in the 5 –3 direction. Thus, the second strand is synthesised discontinuously, in short bursts, about 1000–2000 nucleotides at a time. These short stretches of DNA are called Okazakifragments, after their discoverers.On the lagging strand, a new RNA primer is needed at the start of every Okazaki fragment. These short sequences of RNA are later removed by DNA polymerase I, which
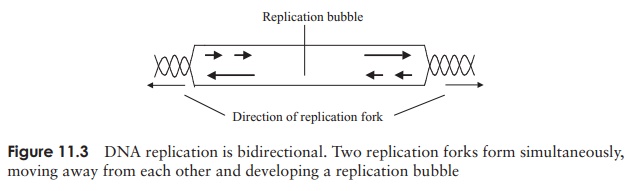
then replaces them with DNA nucleotides. Finally, the fragments are joined together by the action of DNAligase. Replication is bidirectional (Figure 11.3), withtwo forks moving in opposite directions; when they meet, the whole chromosome is copied and replication is complete* .
Related Topics