Chapter: Essential Microbiology: Microbial Genetics
Molecular basis of mutations
The molecular basis of mutations
Any alteration made to the DNA sequence of an organism is called a mutation. This may or may not have an effect on the phenotype (physically manifested properties) of the organism. It may for example enable bacteria to grow without the need for a particular growth supplement, or confer resistance to an antibiotic. Just how this happened was for many years a source of debate. Since the mutant forms only became apparent after the change in conditions (e.g. withdrawal of a nutrient, addition of antibiotic), have some of the bacteria been induced to adapt to the new conditions, or are mutant forms arising all the time at a very low frequency, and merely selected by the environmental change? In 1943, Salvador Luria and Max Delbruck¨ devised the fluctuation test to settle the matter. Results of the fluctuation test together with other evidence led to the understanding that mutations occur spontaneously in nature at a very low frequency. As we shall see later on in this section, however, they can also be induced by a variety of chemical and physical agents. Any change to the DNA sequence is heritable, thus mutations represent a major source of evolutionary variation. Bacteria make marvellous tools for the study of mutations because of their huge numbers and very short generation times.
Since the DNA sequence of a gene represents highly ordered coded information, most mutations have a neutral or detrimental effect on the organism’s phenotype, but occasionally a mutation occurs which confers an advantage to an organism, making it better able to survive and reproduce in a particular environment. Mutants that are favoured in this way may eventually become the dominant type in a population, and, by steps like this, evolution gradually takes place.
Mutations occur spontaneously in any part of an organism’s genome. Spontaneous mutations causing an inactivation of gene function occur in bacteria at the rate of about one in a million for a given gene at each round of cell division. Most genes within a given organism show similar rates of mutation, relative to their gene size; clearly a larger ‘target’ will be ‘hit’ more often than a small one.
How do mutations occur?
Figure 11.17(a) reminds us how the code in DNA is transcribed into messenger RNA and then translated into a sequence of amino acids. Each time the DNA undergoes repli-cation, this same sequence will be passed on, coding for the same sequence of amino acids. Occasionally, mistakes occur during replication. Cells have repair mechanisms to minimise these errors, but what happens if a mistake still slips through? In Figure 11.17(b), we can see the effect of one nucleotide being inserted into the strand instead of another. When the next round of replication occurs, the modified DNA will act as a template for a newly synthesised strand, which at this position will be made comple-mentary to the new, ‘wrong’ base, instead of the original one, and thus the mistake will be perpetuated.


This is an example of the simplest type of mutation, a point mutation, where one nucleotide has been substi-tuted by another. The example shown is a missense muta-tion, which has resulted in the affected triplet coding for a different amino acid; this may or may not have an ef-fect on the phenotype of the organism. RNA polymerase, which transcribes the DNA sequence into mRNA, is un-able to tell that an error has occurred, and faithfully
The machinery of translation is similarly ‘unaware’ of the mistake, and as a consequence, a different amino acid will be inserted into the polypeptide chain. The consequence of expressing a ‘wrong’ amino acid in the protein product could range from no effect at all to a total loss of its biological properties. This can be understood in terms of protein structure, and depends on whether the amino acid affected has a critical role (such as part of the active site of an enzyme), and whether the replacement amino acid has similar or different polar/non-polar properties. You may recall that the genetic code is degenerate, and that most amino acids are coded for by more than one triplet; this means that some mutations do not affect the amino acid produced; such mutations are said to be silent, as in Figure 11.17(c). These most commonly occur at the third nucleotide of a triplet.
Another type of point mutation is a nonsense muta-tion. Remember that of the 64 possible triplet permuta-tions of the four DNA bases, three are ‘stop’ codons, which terminate a polypeptide chain. If a triplet is changed from a coding to a ‘stop’ codon as shown in Figure 11.17(d), then instead of the whole coding se-quence being read, translation will end at this point, and a truncated (and probably non-functional) protein will result.
Mutations can add or remove nucleotides
Other mutations involve the insertion or deletion of nucleotides. This may involve anything from a single nucleotide up to millions. Deletions occur as a result of thereplication machinery somehow ‘skipping’ one or more nucleotides. If the deletion is a single nucleotide, or any-thing other than a multiple of three, the ribosome will be thrown out of its correct reading frame, and a completely new set of triplet codons will be read (Figure 11.18). This is known as a frameshift mutation, and will in most cases result in catastrophic changes to the final protein prod-uct. If the deletion is a multiple of three nucleotides, the reading frame will be preserved and the effect on the protein less drastic.
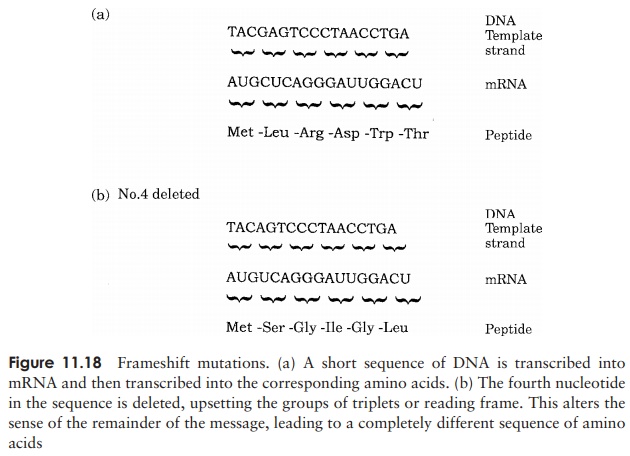
Mutations can be reversed
Just as it is possible for a mutation to occur spontaneously, so it is possible for the nucleotide change causing it to be spontaneously reversed – in other words, a mutant can mutate back to being a wildtype. This is known for obvious reasons as a reverse or back mutation. When this happens, the original genotype and phenotype are restored. Whereas a forward mutation results from any change that inactivates a gene, a back mutation is more specific; it must restore function to a protein damaged by a specific mutation. Not surprisingly, given this specificity, the rate of back mutations is much less frequent.
It is possible for the wildtype phenotype to be restored, not through a reversal of the original base change, but due to a second mutation at a different location. The effect of this second mutation is to suppress the effects of the first one. These are called suppressor or second site mutations. They are double mutants that produce apseudowildtype; the phenotype appears to be wildtype, but the genotype differs.
Mutations have a variety of mechanisms
Why are mistakes made every so often during DNA replication? One source of erro-neous base incorporation is a phenomenon called tautomerism. Some of the nucleotide bases occur in rare alternative forms, which have different base-pairing properties. For example, as you will recall, cytosine normally has an amino group that provides a hydrogen atom for bonding with the complementary keto group of gua-nine. Occasionally, however (once in about every 104–105 molecules), the cytosine may undergo a rearrangement called a tautomeric shift, which results in the amino group changing to an imino group (==NH), and this now behaves in pairing terms as if it were thymine, and therefore pairs with adenine (Figure 11.19a). Similarly, thymine might undergo a tautomeric shift, changing its usual keto form (C==O) to the rare enol form (COH). This then takes on the pairing properties of cytosine and pairs with guanine (Fig-ure 11.19b). The result of such mispairing is that at subsequent rounds of replication,
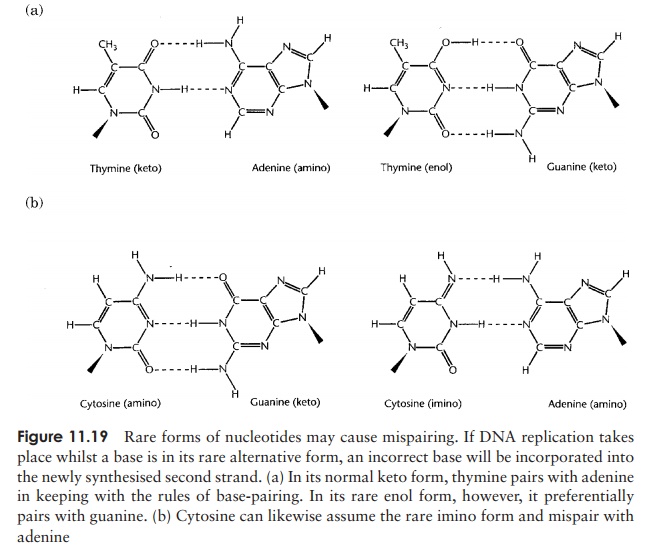
one half of the DNA molecules will contain a wrong base pair at that point. The fact that spontaneous mutations occur much less frequently than the rate just quoted is due to the DNA repair mechanisms.
Mutations also occur in viruses
As we saw, the genome of viruses can be composed of either DNA or RNA, and these are subject to mutation just like cellular genomes. The rate of mutation in RNA viruses is much higher than in those containing DNA, about a thousand times higher, in fact. This is significant, because by frequently changing in this way, pathogenic viruses are able to stay one step ahead of host defence systems.
Mutagenic agents increase the rate of mutations
In the early part of the 20th century, the existence of mutations was appreciated, but attempts to gain a fuller understanding of them were hampered by the fact that they occurred so rarely. In the 1920s, however, it was shown that the mutation rate in both barley plants and the fruit fly Drosophila was greatly increased as a result of exposure to X-rays. In the following decades, a number of chemical and physical agents were shown also to cause mutations. Mutagens such as these raise the general level of mutations in a population, ratherthan the incidence of mutation in at particular locationin the genome. They are extremely useful tools for themicrobial geneticist, but need to be treated with greatcare because they are also mutagenic (and in most casescarcinogenic) towards humans.Chemical mutagens can be divided into five classes:
Base analogues
A base analogue is able to ‘mimic’ one of the four normal DNA bases by having a chemical structure sufficiently similar for it to be incorporated into a DNA molecule instead of that base during replication. One such base is 5-bromouracil (5-BU); in its usual keto form, it acts like thymine (which it closely resembles – see Figure 11.20), and therefore pairs with adenine, and is not mutagenic. However 5-BU is capable of tau-tomerising to the enol form, which, remember, pairs with G, not A. Whereas for thymine the enol form is a rarity, for 5-BU it is more common, hence mispairing with guanine occurs more frequently. So, we get the same outcome as with spontaneous mutation – the TA has been replaced by a CG – but its frequency is much increased. Several other base analogues, such as 2-aminopurine, which mimics adenine, have similar effects to 5-BU. Mutations brought about by base analogues all result in a purine being replaced by another purine, or a pyrimidine being replaced by another pyrimidine; this type of mutation is called a transition.
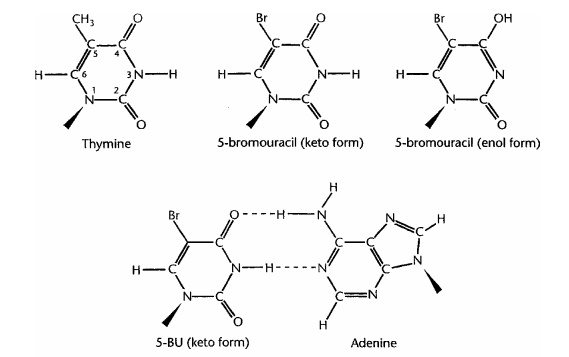
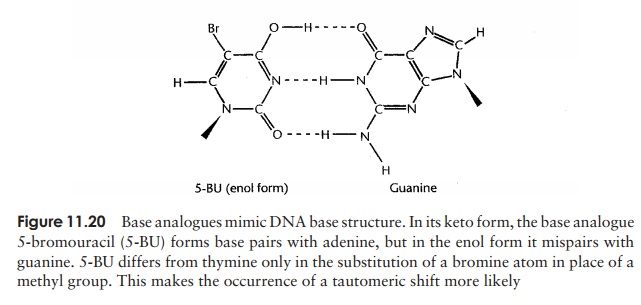
Alkylating agents
As the name suggests, this group of mutagens act by adding an alkyl group (e.g. methyl, ethyl) at various positions on DNA bases. Ethylethanesulphonate (EES) and ethyl-methanesulphonate (EMS) are alkylating agents, used as laboratory mutagens. They act by alkylating guanine or thymine at the oxygen atom involved in hydrogen bond-ing. This leads to an impairment of the normal base-pairing properties, and causes guanine for example to mispair with thymine.
Substitutions brought about by alkylating agents can be either transitions or trans-versions.
Deaminating agents
Nitrous acid is a potent mutagen, which alters the base-pairing affinities of cytosine and adenine by replacing an amino group with an oxygen atom. Transitions occur in both directions, AT → GC and GC → AT. Note that although guanine is deaminated, its base pairing properties are not affected.
Intercalating agents
These mutagens exert their effect by inserting themselves between adjacent nucleotides in a single strand of DNA. They distort the strand at this site and cause either the addition or, less commonly, deletion of a nucleotide.
This leads to frameshift mutations, and often during replication the bound mutagen interferes with new strand synthesis, leading to a gross deletion of nucleotides. Ethidium bromide, commonly used for the visualisation of small amounts of DNA in the molecular biology laboratory, is an intercalating agent; it has a planar structure with roughly the same dimensions as a purine–pyrimidine pairing.
Hydroxylating agents
Hydroxylamine has a specific mutagenic effect, hydroxylating the amino group of cy-tosine to cause the transition of GC → AT.
Base analogues and certain intercalating agents can only exert their mutagenic effects if they are incorporated into DNA while it is replicating. Others, which depend on altering base pairing by modifying the structure of DNA bases, are effective on both replicating and non-replicating DNA.
Physical mutagens
As mentioned at the start of this section, the first mutagenic agent to be demonstrated were X-rays; along with ultraviolet light, they are the most commonly used physical mutagen. X-rays, like other forms of ionising radiation, cause the formation of highly reactive free radicals. These can bring about changes in base structure as well as gross chromosomal alterations. DNA strands can be broken and reannealed incorrectly, to produce errors in the sequence.
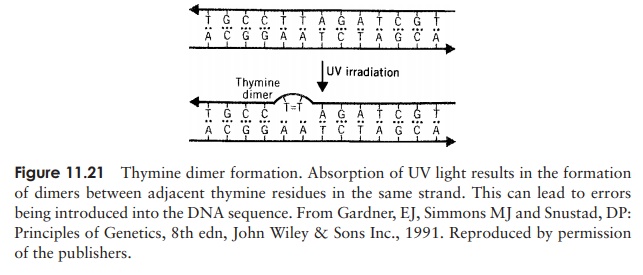
In recent years, we have all become much more aware of the possible perils of ex-cessive exposure to sunlight. Ultraviolet light (UV) from the sun damages DNA in skin cells, which can lead to them becoming cancerous. The specific action is on adjacent pyrimidine bases (usually thymines) on the same strand, which are cross-linked to form a dimer (Figure 11.21). This results in a distortion of the helix and interferes with replica-tion. UV light is most effective at wavelengths around 260 nm, as this is the wavelength most strongly absorbed by the DNA bases.
DNA damage can be repaired
All organisms have developed the means to repair damage to their DNA, in addition to the proofreading mechanism described earlier
The most common way of dealing with mutations is by means of a method called excision repair, in which enzymes recognise and cut out the altered region of DNAand then fill in the missing bases, using the other strand as a template (Figure 11.22). Mismatch repair is used to repair the incorporation of a base that has escaped theproofreading system. In a situation such as this, it is not immediately obvious which is the correct base and which is the mistake, so how does the cell know which strand to replace? In E. coli, the old strand is distinguished from the new by the fact that some of its bases are methylated. This only occurs some time after replication, so newly synthesised strands will not have the methyl groups and can thus be recognised.
Less frequently, the alteration in DNA structure is simply reversed by direct repair mechanisms, the best known of which is photoreactivation. This involves an enzyme
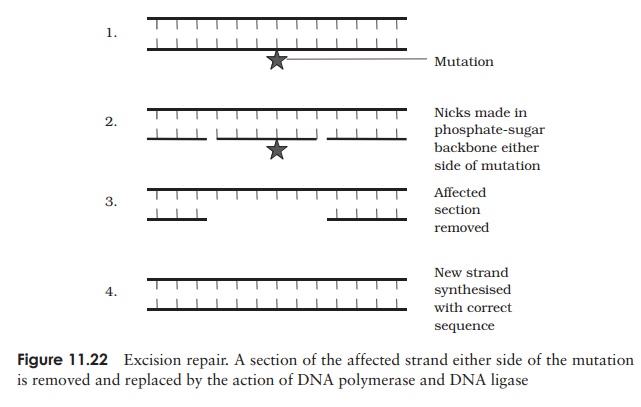
calledDNA photolyase, which breaks the bonds formed between adjacent thymine bases by UV light before replication takes place. Unusually, the photolyase is dependant on visible light (>300 nm) for activation. Another form of direct repair, involving the enzyme methylguanine transferase allows the reversal of the effects of alkylating agents.
Loss of repair mechanisms can allow mutations to become established which would normally be corrected. A harmful strain of E. coli which emerged in the 1980s was shown to have developed its pathogenicity due to a deficiency in its repair enzymes.
Carcinogenicity testing: the Ames test
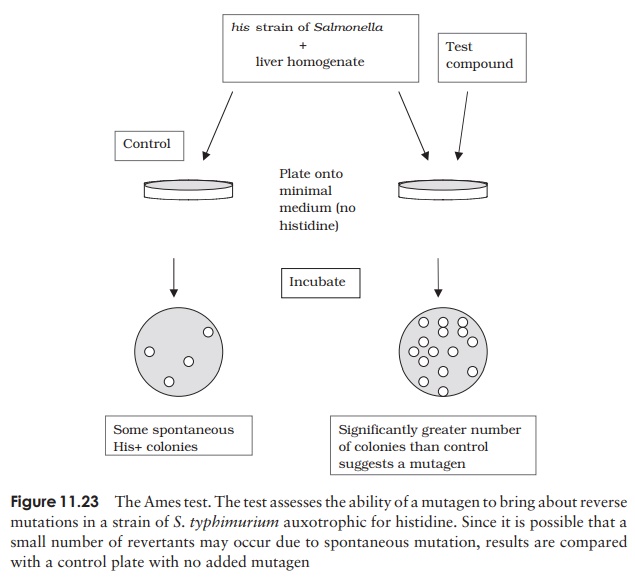
The great majority of carcinogenic substances, that is, substances that cause cancer in humans and animals, are also mutagenic in bacteria. This fact has been used to develop an initial screening procedure for carcinogens; instead of the expensive and time-consuming process of exposing laboratory animals (not to mention the moral issues involved), a substance can be tested on bacteria to see if it induces mutations.
The Ames Test assesses the ability of a substance to cause reverse mutations in aux-otrophic strains of Salmonella that have lost the ability to synthesise the amino acid his-tidine (his− ). Rates of back mutation (assessed by the ability to grow in a histidine-free medium) are compared in the presence and absence of the test substance (Figure 11.23). A reversion to his+ at a rate higher than that of the control indicates a mutagen. Many substances are procarcinogens, only becoming mutagenic/carcinogenic after metabolic conversion by mammals; in order to test for these, an extract of rat liver is added to the experimental system as a source of the necessary enzymes.
Related Topics