Chapter: Essential Microbiology: Microbial Genetics
Regulation of gene expression
Regulation of gene expression
The proteins synthesised by microorganisms may broadly be divided into structural proteins required for the fabric of the cell, and enzymes, used to maintain the essential metabolic processes of the cell. It would be terribly wasteful (and possibly harmful) to produce all these proteins incessantly, regardless of whether or not the cell actually re-quired them, so microorganisms, in common with other living things, have mechanisms of gene regulation, whereby genes can be switched on and off according to the cell’s requirements.
Induction of gene expression
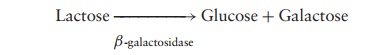
The synthesis of many enzymes required for the catabolism (breakdown) of a substrate is regulated by en-zyme induction. If the substrate molecule is not present in the environment, there is little point in synthesising the enzyme needed to break it down. In terms of conser-vation of cellular energy, it makes more sense for such enzymes to be produced only when they are needed, that is, when the appropriate substrate molecule is present. The substrate itself therefore acts as the inducer of the en-zyme’s synthesis. An example of this that has been stud-ied in great depth concerns the enzyme β-galactosidase, used by E. coli to convert the disaccharide lactose into its constituent sugars:
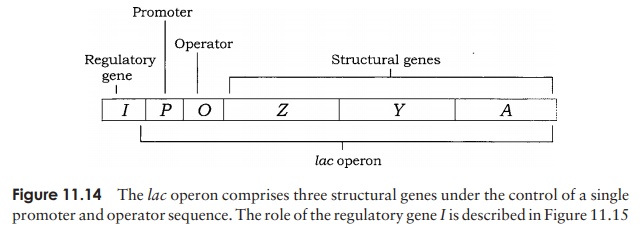
β-galactosidase has been intensively studied as part of the lac operon of E. coli. This is made up of three structural genes designated Z, Y and A, which are clus-tered together and share a common promoter and ter-minator (Figure 11.14). The genes code for, respectively, β-galactosidase, a permease and a transacetylase. The permease is necessary for the transport of lactose into the cell, while the role of the transacetylase is not en-tirely clear, although it is essential for the metabolism of lactose. Grouping the three genes together in this way en-sures an ‘all-or-nothing’ expression of the three proteins. Transcription of these structural genes into their re-spective mRNAs is initiated by the enzyme RNA poly-merase binding to the promoter sequence. However, this is only possible in the presence of lactose; in its ab-sence, a repressor proteinbinds to an operator site, adja-cent to the promoter, preventing RNA polymerase bind-ing to the promoter, and therefore preventing mRNA production. Production of the repressor protein is encoded by a regulator gene (I), situated slightly upstream from the operon (Figure 11.15a).
How then, does the presence of lactose overcome this regulatory mechanism? Allo-lactose, an isomer of lactose and an intermediate in its breakdown, attaches to a site on the lac repressor, thereby reducing the latter’s affinity for the operator, and neutral-ising its blocking effect (Figure 11.15b). The structural genes are then transcribed into mRNA, which is subsequently translated into the three proteins described above, and the lactose is broken down. In the absence of lactose, there are only trace amounts ofβ-galactosidase present in anE. colicell; this increases some 1000-fold in its presence.
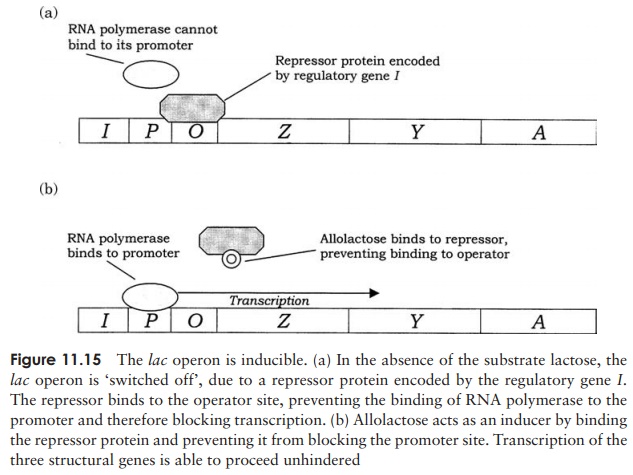
When all the lactose has been consumed, the repressor protein is free to block the operator gene once more, and the needless synthesis of further β-galactosidase ceases.
The lac operon can also be induced by isopropyl β-thiogalactoside (IPTG); E. coli is not able to break this down, so the genes remain permanently switched on. IPTG is utilised as an inducer in cloning systems involving the expression of the lacZ gene on pUC plasmids.
The lac operon is subject to control by positive as well as negative regulator proteins. Transcription of the operon only occurs if another regulatory protein calledcataboliteactivator protein (CAP) is bound to the promoter sequence. This isdependent on a relatively high concentration of the nucleotide cAMP which only occurs when glucose is scarce. The activation of the lac operon thus occurs only if lactose is present and glucose is (almost) absent.
Repression of gene expression
The induction of gene expression, such as we have just described for the lac operon, generally relates to catabolic (breakdown) reactions. Anabolic (synthetic) reactions, such as those leading to the production of specific amino acids, by contrast, are often controlled by the repression of key genes.
Enzyme repression mechanisms operate along similar lines to induction mechanisms, but the determining factor here is not the substrate of the enzymes in question (lactose in our example), but the end-product of their action. The trp operon contains a cluster of genes encoding five enzymes involved in the synthesis of the amino acid tryptophan. (Figure 11.16) In the presence of tryptophan, the cell has no need to synthesise its
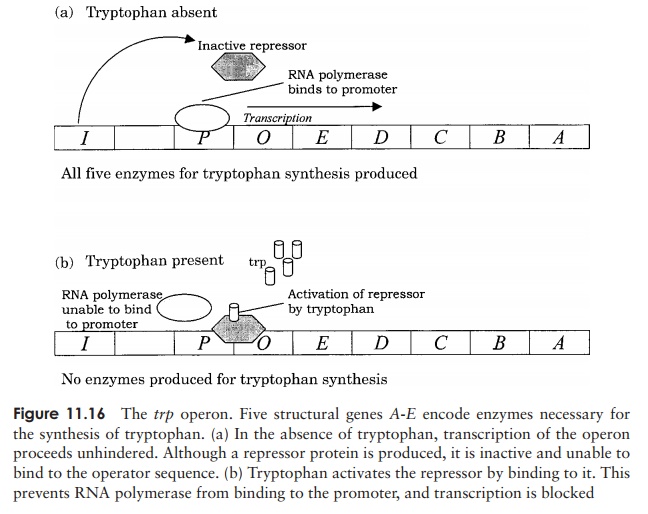
own, so the operon is switched off. This is achieved by tryptophan binding to and activating a repressor protein, which in turn binds to the operator of the trp operon and prevents transcription of the synthetic enzymes. The tryptophan here is said to act as a corepressor. As tryptophan is used up and its level in the cell falls, the repressor revertsto its inactive form, allowing transcription of the tryptophan-synthesising enzymes to go ahead unhindered.
Global gene regulation
The systems of gene regulation discussed above apply to individual operons; sometimes, however, a change in environmental conditions necessitates the regulation of many genes at once. These global regulatory systems respond to stimuli such as oxygen depletion and temperature change, and utilise a number of different mechanisms.
Related Topics