Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Bacterial Genetics
Transposable Elements - Bacterial Genetics
TRANSPOSABLE ELEMENTS
Transposable elements are genetic units that are capable of mediating their own transferfrom one chromosome to another, from one location to another on the same chromosome, or between chromosome and plasmid. This transposition relies on their ability to synthe-size their own site-specific recombination enzymes, called transposases.
The three major kinds of transposable elements are insertion sequence elements; transposons; and certain prophages, such as Mu.
Insertion Sequence Elements
Insertion sequence (IS) elements are segments of DNA of approximately 1000 bp. Theyencode enzymes for site-specific recombination and have distinctive nucleotide sequences at their termini. Different IS elements have different termini, but, as illustrated in Figure 4 – 8, a given IS element has the same sequence of nucleotides at each end, but in an inverted order. Only genes involved in transposition (eg, one encoding a transposase) and in the regulation of its frequency are included in IS elements, and they are therefore the simplest transposable elements.
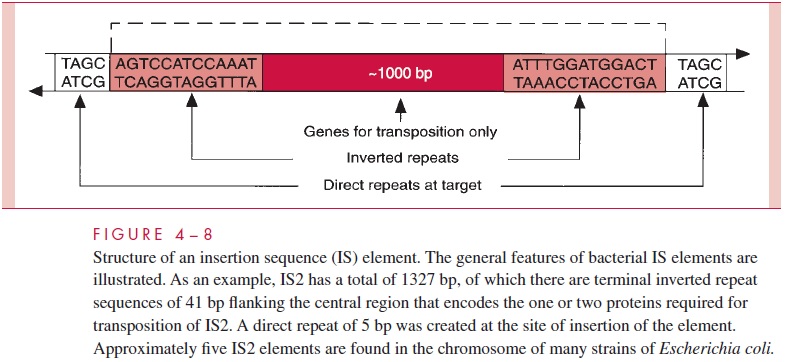
Because IS elements contain only genes for transposition, their presence in a chro-mosome is not always easy to detect. However, if an IS element transposes to a new site that is within a gene, this insertion is actually a mutation that alters or destroys the activ-ity of the gene. Because most IS elements contain a transcription termination signal, the insertion also eliminates transcription of any genes downstream in the same operon. This property of IS elements led to their first recognition. Reversion of insertion mutations can occur by deletion, but the frequency of deletion is 100- to 1000-fold lower than that of insertion.
Numerous IS elements reside naturally at different locations in E. coli chromosomes and in E. coli plasmids, and this has many consequences for the cell. Because their size is sufficient to permit strong base pairing between different copies of the same IS element, they can provide the basis for RecA-mediated homologous recombination. In this man-ner, the presence of particular IS elements in both the F plasmid and the bacterial chro-mosome provides a means for the formation of Hfr molecules by cointegration using IS sequence homology and the RecA system.
Transposons
One of the major aspects of IS elements is that they are components of transposons (Tn elements), which are transposable segments of DNA containing genes beyond those needed for transposition. Transposons are as much as 10-fold larger than IS elements. One class, of which transposon Tn10 is a good example, are composite structures consist-ing of a central area of genes bordered by IS elements. The genes may code for such properties as antimicrobic resistance, substrate metabolism, or other functions. A general-ized transposon structure of the Tn10 variety is shown in Figure 4 – 9.
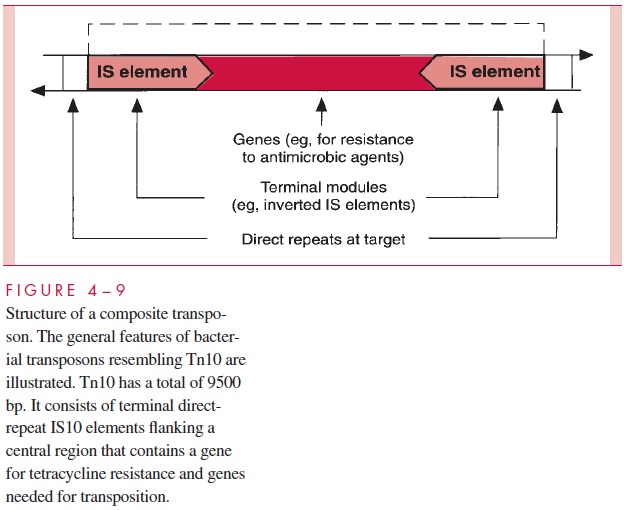
Composite transposons of the Tn10 sort can translocate by what is called simple or direct transposition, in which the transposon is excised from its original location and in-serted without replication into its new site. A second class, typified by transposon Tn3, has inverted repeat sequences rather than IS elements at its ends and encodes not only a transposase but also an enzyme called a resolvase. Transposition of Tn3 involves forma-tion of acointegrate of the two DNA molecules (or segments of the same molecule) in-volved in the transposition — that is, the one carrying the Tn3 and the one serving as the target. Replication of the transposon then occurs, and the resolvase separates (resolves) the cointegrate, restoring the two DNA molecules, each now with its own copy of Tn3. Transposition of this sort is called replicative or duplicative transposition.
Besides the primary insertion reaction, all transposable units promote other types of DNA rearrangements, including deletion of sequences adjacent to a transposon, inversion of DNA segments, fusion of separate plasmids within a cell, similar fusions that integrate plasmids with the cell chromosome, and repeated duplications that result in amplification of genes within transposons. All of these events have great significance for understanding the formation and spread of antimicrobic resistance through natural populations of path-ogenic organisms. These subjects are discussed in the description of plasmids in the next section.
Some strains of streptococci harbor transposon-like, drug-resistance elements within their chromosome that are capable of mediating their own transfer to other cells by con-jugation. One such conjugative transposon is Tn916, found originally in a strain of E. faecalis. This element, approximately 16 kb in size, contains a gene for tetracyclineresistance. It and similar elements resemble transposons in many respects, including size, multiple target sites, ability to transfer from a chromosome to a plasmid, and ability to be removed from a plasmid or a chromosome by precise excision. What is unusual, however, is their ability to mediate their own intercellular transfer. It now appears that Tn916, and presumably similar elements, can form a transient plasmid-like structure as part of the process of conjugational transfer.
The third type of transposable element is transposable prophage, such as that of bac-teriophage Mu, which has the alternative of lytic growth or of lysogeny. During lysogeny, the prophage of Mu can insert virtually anywhere in the E. coli chromosome and later can transpose itself from one location to another. In fact, it is a transposon. When it integrates within a bacterial gene, it inactivates it in the same manner as any other transposable ele-ment. It was originally recognized as a virus that causes mutation, hence its name.
Invertible DNA Segments and Recombinational Regulation of Gene Expression
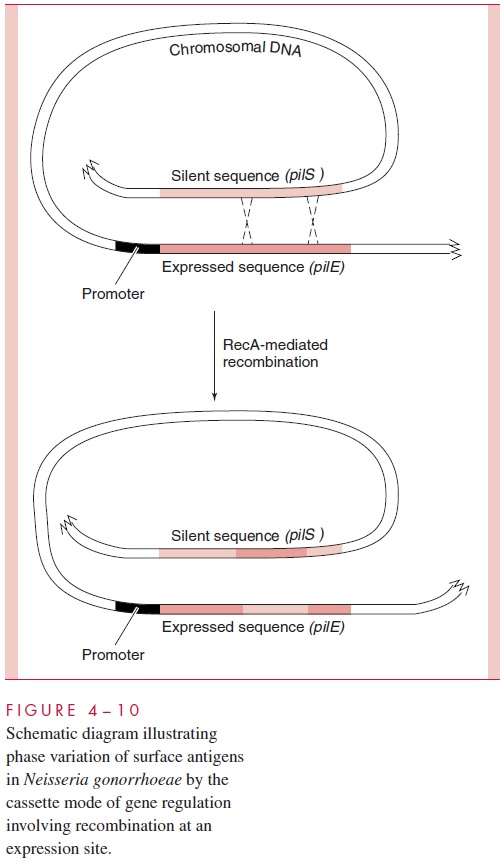
A fascinating aspect of DNA rearrangements brought about by genetic recombination is that the expression of some chromosomal genes important in virulence are actually con-trolled by recombinational events. All the known cases involve phase variation of surface antigens. In N. gonorrhoeae, the bacteria that causes gonorrhea , multiple genes encoding antigenically different pilin sequences exist throughout the chromosome. Many, called pilS, are silent because they lack effective promoters; some are only frag-ments of pilin sequences. These silent genes or gene fragments serve as a reservoir of anti-genic variability; each can, wholly or in part, become inserted by RecA-dependent homol-ogous recombination into an actively expressed gene (pilE), resulting in the synthesis of a new pilin. The entire process resembles the insertion of cassette tapes into a tape player and, therefore, is referred to as the cassette mode of gene regulation (Fig 4 – 10).
A different DNA rearrangement is responsible for the alternation of expression of anti-genically distinct flagellins, H1 and H2, in Salmonellaspecies. An invertible element of 995 bp lies between the two flagellin genes (Fig 4 – 11). The phase-2 encoding gene (B) lies in an operon that also encodes a repressor for the phase-1 encoding gene (C). The latter gene is, therefore, active only if the former operon is inactive. Activity of the phase-2 operon, which lacks its own promoter, depends on a promoter within the invertible element.
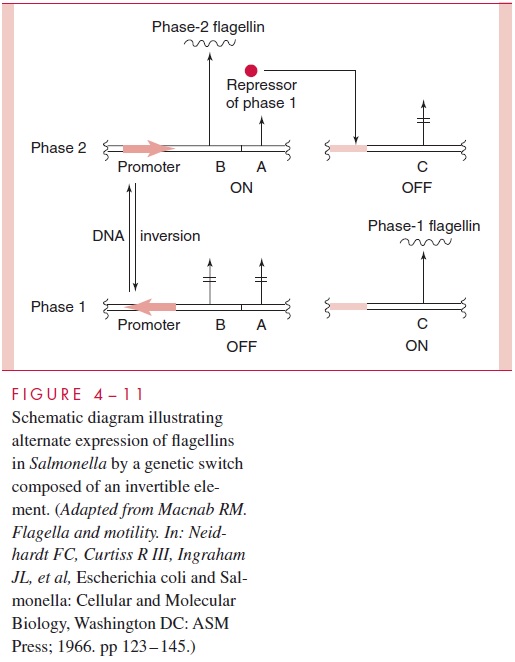
In one orienta-tion, this promoter can initiate transcription of the B gene; in the other orientation transcrip-tion, if it starts, proceeds in the opposite direction, and the B gene is silent, allowing the C gene to work. In this manner, excision of the invertible element and its reinsertion at the same site but in the opposite orientation lead to a shift from one flagellar form to the other (ie, to antigenic phase variation). The invertible element encodes its own site-specific recombinaseenzyme that catalyzes the inversion in response to currently unknown signals. A similar situ-ation exists in E. coli, where a 314-bp invertible segment containing a promoter controls tran-scription of the adjacent, promoter-less fimA gene. This gene encodes the structural protein for type 1 (common) pili, which function as an adhesin in mediating the binding of E. coli to eukaryotic cells, thereby aiding in the early stages of tissue colonization by these bacteria.
It is believed that antigenic variation mediated by these site-specific transpositional rearrangements provides a selective advantage to the bacteria in allowing invading popu-lations to include individuals that can escape the developing immune response of the host and thus continue the infectious process. Similar strategies are used by some eukaryotic parasites of humans, notably the trypanosomes .
Related Topics