Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Bacterial Genetics
More about Bacterial Plasmids
MORE ABOUT BACTERIAL PLASMIDS
One of the unanticipated features of microbial genetics has been the revelation that many virulence factors and much clinically significant resistance to antibiotics are the result of the activities not of bacterial chromosomal genes but of the accessory genomes present in plasmids. In a certain sense, the health professional treating infectious disease is fre-quently coping with autonomous self-replicating DNA molecules. Many of the properties of plasmids have already been touched on, but the information is now consolidated and considered in more detail.
General Properties and Varieties of Plasmids
We have already encountered plasmids in our consideration of conjugation. To recap, plas-mids are ubiquitous extrachromosomal elements composed of double-stranded DNA that typically is circular (Fig 4 – 12). (Linear plasmids occur in medically relevant strains of Borrelia.) A single organism can harbor several distinct plasmids. Like the chromosome,they have the property of governing their own replication by means of special sequences and regulatory proteins, including a genetic region called ori (origin of replication) at which specific proteins initiate replication. Any DNA molecule that is self-reproducing, in-cluding all plasmids as well as the bacterial chromosome, is said to be a replicon.
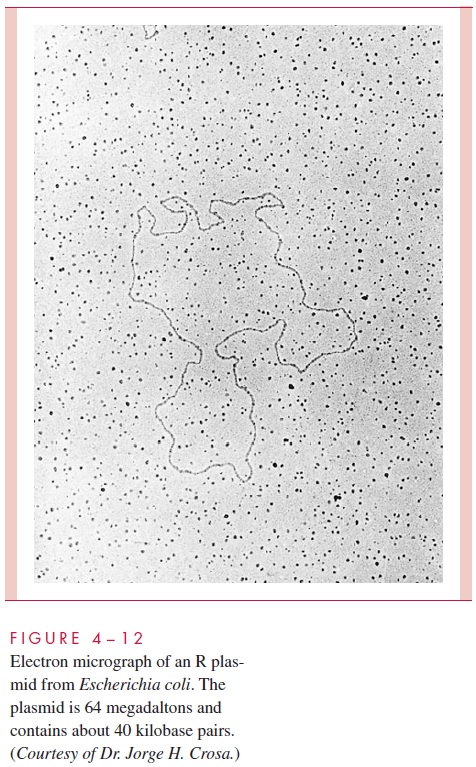
Plasmids vary greatly in size, in the mode of control of their replication, and in the number and kinds of genes they carry. Naturally occurring plasmids range from less than 5 million to more than 100 million daltons, but even the largest are only a few percent of the size of the bacterial chromosome.
The number of molecules of a given plasmid that is present in a cell, called the copy number, varies greatly among different plasmids, from only a few molecules per cell to dozens of molecules per cell. In general, small plasmids tend to be represented by more copies per cell.
Conjugal transfer is an important property of those plasmids that possess the com-plex of tra genes, but even nonconjugative plasmids can transfer to some extent to other cells as a result of mobilization by conjugative plasmids. Some plasmids, again including the F factor, can replicate either autonomously or as a segment of DNA inte-grated into the chromosome. These are sometimes termed episomes. Certain prophages can exist as plasmids, but most plasmids are not viruses, because at no point of their life cycle do they exist as a free viral particle (virion). Most plasmids show little or no DNA homology with the chromosome and can, in this sense, be regarded as foreign to the cell.
Plasmids usually include a number of genes in addition to those required for their replication and transfer to other cells. The variety of cellular properties associated with plasmids is very great and includes fertility (the capacity for gene transfer by conjuga-tion), production of toxins, production of pili and other adhesins, resistance to antimicro-bics and other toxic chemicals, production of bacteriocins (toxic proteins that kill some other bacteria), production of siderophores for scavenging Fe3+ , and production of certain catabolic enzymes important in biodegradation of organic residues.
On the other hand, plasmids can add a small metabolic burden to the cell, and in many cases, a slightly reduced growth rate results. Thus, under conditions of laboratory cultivation where the properties coded by the plasmid are not required, there is a tendency for curing of a strain to occur, because the progeny cells that have not acquired a plasmid (or have lost it) have a selective advantage during prolonged growth and subculture. Con-versely, where the property conferred by the plasmid is advantageous (eg, in the presence of the antimicrobic to which the plasmid determines resistance), selective pressure favors the plasmid-carrying strain.
Although plasmids are central to infectious disease and have been studied for decades, their origin remains uncertain. They could possibly be descendants of bacterial viruses that evolved a sophisticated means of self-transfer by conjugation and then lost their unneeded protein capsid. Alternatively, they may have evolved as separated parts of a bacterial chromosome that could provide both the means for genetic exchange and a way to amplify certain genes of special value in a particular environment (eg, coding for an adhesin) or to dispense with them where they are superfluous.
A great many bacterial plasmids are known. Some show similarity with each other in nucleotide sequence; thus, plasmids can be classified by their degree of apparent related-ness. Unrelated plasmids can coexist within a single cell, but closely related plasmids be-come segregated during cell division and eventually all but one are eliminated. For this reason, a group of closely related plasmids that exclude each other are referred to as anincompatibility group.
R Plasmids
Plasmids that include genes conferring resistance to antimicrobics are of great significance to medicine. They are termed R plasmids or R factors(resistance factors). The genes responsible for resistance usually code for enzymes that inactivate antimicrobics or reduce the cell’s permeability to them. In contrast, resistance conferred by chromosomal mutation usually involves modification of the target of the antimicrobics (eg, RNA polymerase or the ribosome).
R plasmids occupy center stage in approaches to chemotherapy because of the con-stellation of properties they possess. Those of Gram-negative bacteria can be transmitted across species boundaries and, at lower frequency, even between genera. Many encode re-sistance to several antimicrobics and can thus spread multiple resistance through a diverse microbial population under selective pressure of only one of those agents to which they confer resistance. Nonpathogenic bacteria can serve as a natural reservoir of resistance determinants on plasmids that are available for spread to pathogens.
R plasmids evolve rapidly and can easily acquire additional resistance-determining genes from fusion with other plasmids or acquisition of transposons. Many have the capa-bility of amplifying the number of copies of their resistance genes either by gene duplica-tions within each plasmid or by increasing the number of plasmids (copy number) per cell. By these means resistance can be achieved to very high concentrations of the antimi-crobic. One process of gene amplification is based on the ability of some conjugative plasmids to dissociate their components into two plasmids, one (called the resistancetransfer factor) containing genes for replication and for transfer and another (calledthe resistance or r determinant) containing genes for replication and for resistance. Subsequent relaxed replication of the r determinant expands the cell’s capacity to produce the resistance-conferring enzyme (Fig 4 – 13).
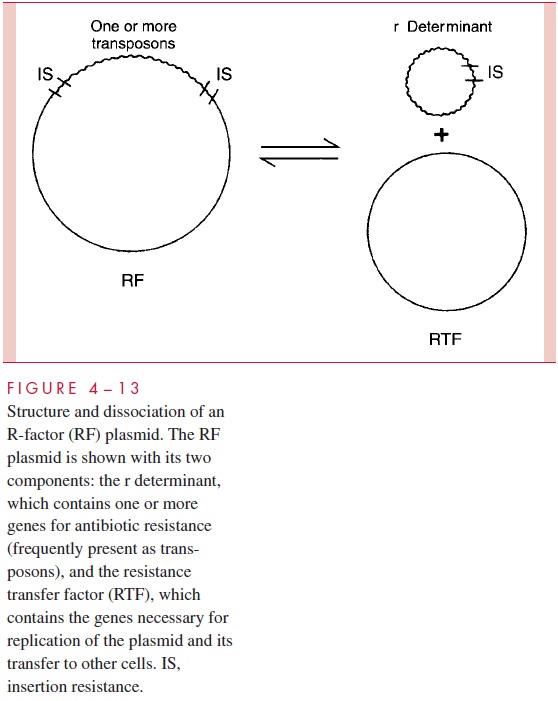
Over the past three decades, many of the molecular feats of R plasmids have been explained on the basis of known genetic and evolutionary mechanisms. The discovery of transposable elements (insertion sequences and transposons) and their properties provides an explanation for many of these phenomena. Most plasmids, and all R factors, contain many IS elements and transposons. In fact, virtually all the resistance determinant genes on plasmids are present as transposons. As a result, these genes can be amplified by tan-dem duplications on the plasmid and can hop to other plasmids (or to the bacterial chro-mosome) in the same cell. Combined with the natural properties of many plasmids to transfer themselves by conjugation (even between dissimilar bacterial species), the rapid evolutionary development of multiple drug resistance plasmids and their spread through populations of pathogenic bacteria during the past three decades can be seen as a pre-dictable result of natural selection resulting from the widespread and intensive use of an-timicrobics in human and veterinary medicine .
The properties of transposons can explain the present-day ubiquity and mobility of resistance genes but not their origin. Two facts help point to at least a direction in which to search for an answer. First, R plasmids carrying the genes encoding antimicrobic-inactivating enzymes have been found in bacterial cultures preserved by lyophilization (freeze-drying) since before the era of antimicrobic therapy; an accelerated evolutionary development need not be invoked. Second, the enzymes themselves are remarkably simi-lar to those found in certain bacteria (Streptomyces spp) that produce many clinically useful antimicrobics. Perhaps a long time ago there was a cross-genus transfer of genetic information (by transformation?) that became stabilized on plasmids under the selection pressure of an antimicrobic released into the environment under natural conditions.
Detection of Plasmids
A number of physical, morphologic, and functional tests can be used to reveal the presence of plasmids in a bacterial population. The rapid transfer of characteristics, such as resis-tance to antimicrobics, from strain to strain or, alternatively, the rapid loss of such traits is a hallmark of plasmid-encoded characteristics. When several genetically distinct character-istics are transferred simultaneously in the laboratory into cells known not to have pos-sessed them previously, the evidence is very strong that a plasmid is responsible. Plasmids, including nonconjugative plasmids and those coding for no presently known trait, can be demonstrated directly by agarose gel electrophoresis. Electron microscopy can also be used to visualize plasmids, to measure the length of their DNA, and to see the forms they take on hybridiza-tion to other nucleic acid molecules (see Fig 4 – 12).
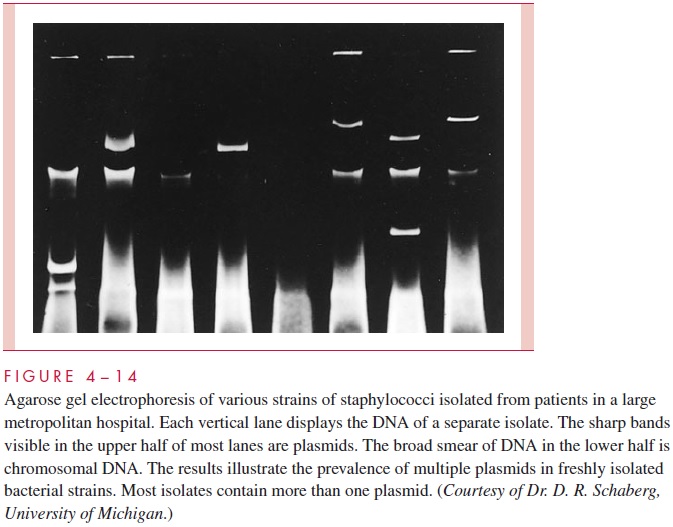
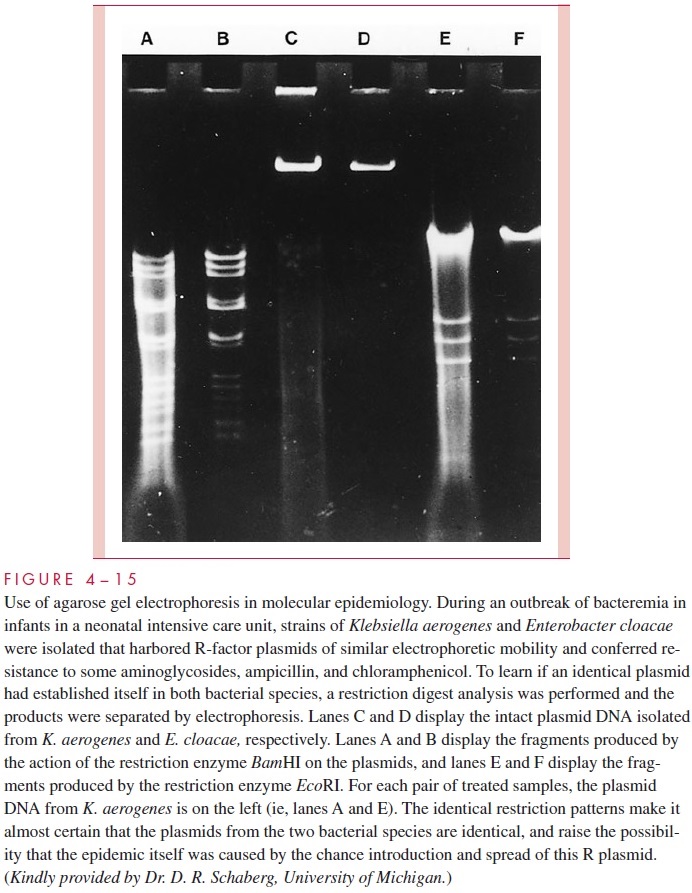
Bacterial plasmids, including R factors, have become valuable markers for comparing closely related strains of bacteria in epidemiologic studies. In outbreaks, spread of an epi-demic strain can sometimes be followed more easily and more accurately by monitoring the profile of plasmids carried in strains isolated from different patients than by using tra-ditional typing methods (Fig 4 – 14). This approach is particularly useful in studying out-breaks of nosocomial (hospital-acquired) infections. Likewise, the spread of an R plasmid between different species can be followed by showing that they carry an identical plasmid conferring the same pattern of antimicrobic resistance. The plasmid comparison can be carried one step further in specificity by cutting the plasmid DNA with specific restriction endonucleases (see next section) and examining the resulting fragments by agarose gel electrophoresis (Fig 4 – 15). Variations of this procedure enable even the spread of specific genes among a variety of plasmids to be detected.
Related Topics