Chapter: Biotechnology Applying the Genetic Revolution: Recombinant Proteins
Translation Expression Vectors
TRANSLATION
EXPRESSION VECTORS
As discussed in Previews
Pages, bacterial ribosomes bind mRNA by recognizing the ribosome binding site
(RBS) (also known as the Shine-Dalgarno sequence). The RBS base pairs with the
sequence AUUCCUCC on the 16S rRNA of the small subunit of the ribosome. The
closer the RBS is to the consensus sequence (i.e., UAAGGAGG), the stronger the
association. Generally, this leads to more efficient initiation of translation.
In addition, for optimal translation, the RBS must be located at the correct
distance from the start codon, AUG.
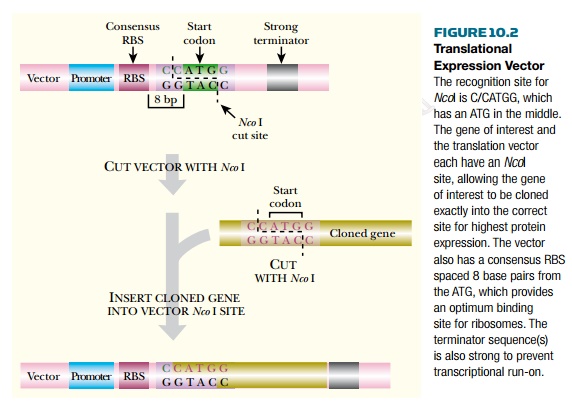
Expression vectors are
designed to optimize gene expression at the level of transcription. However, it
is also possible to design translational
expression vectors to maximize the
initiation of translation. These vectors possess a consensus RBS plus an ATG
start codon located an optimum distance (8 bp) downstream of the RBS. The
cloned gene is inserted into a cloning site that overlaps the start codon. The
restriction enzyme NcoI is very
convenient because its recognition site (C/CATGG) includes ATG. Therefore it is
possible to insert a cloned gene so that its ATG coincides exactly with the ATG
of the translational expression vector (Fig. 10.2). The gene to be expressed is
cut with NcoI at its 5’-end and with
another convenient restriction enzyme at its 3’-end. If necessary, an
artificial NcoI site may be
introduced into the gene by site-directed mutagenesis or by PCR.
Translational expression
vectors also possess a convenient selective marker, usually resistance to
ampicillin or some other antibiotic, and a strong, regulated transcription
promoter. Downstream of the cloning site are two or three strong terminator
sequences, to prevent transcription continuing into plasmid genes.
Although translational expression vectors can optimize the initiation of translation, they do not control the other factors listed earlier, which are properties of the sequence of each individual gene. The secondary structure of the mRNA may have significant effects on the level of translation. If the mRNA folds up so that the RBS and/or start codon are blocked, then translation will be hindered. In particular, the sequence of the first few codons of the coding sequence should be checked for possible base pairing with the region around the RBS. If necessary, bases in the third (redundant) position of each codon may be changed to eliminate such base pairing. Getting active and abundant translation is the key to preventing mRNA instability. Any mRNA that is not being actively translated becomes subject to degradation, which decreases the protein yield.
Related Topics