Chapter: Biotechnology Applying the Genetic Revolution: Recombinant Proteins
Codon Usage Effects
CODON
USAGE EFFECTS
When genes from one organism
are expressed in different host cells the problem of codon usage arises. As
explained in Previews Pages, the genetic code is redundant in the sense that several different codons can encode
the same amino acid. So even though a protein has a fixed amino acid sequence,
there is still considerable choice over which codons to use. In practice,
different organisms favor different codons for the same amino acid. For
example, the amino acid lysine is encoded by AAA or AAG. In E. coli, AAA is used 75% of the time and
AAG only 25%. In contrast, Rhodobacter
does the exact opposite and uses AAG 75% of the time, even though E. coli and Rhodobacter are both gram-negative bacteria.
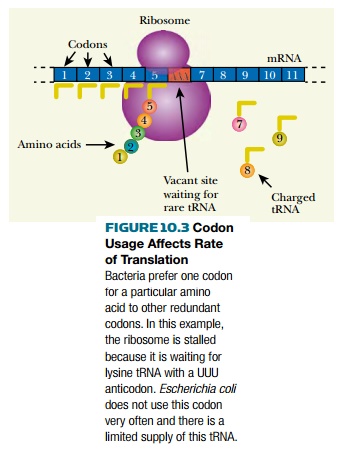
Codons are read by transfer
RNA. When a cell uses a particular codon only rarely, it has lower levels of
the tRNA that reads the rare codon. So if a gene with many AAA codons is put
into a cell that rarely uses AAA for lysine, the corresponding tRNA may be in
such short supply that protein synthesis slows down (Fig. 10.3).
Consequently, to optimize
protein production, codon usage must be considered. Although it takes a lot of
work, genes may be altered in DNA sequence so as to change many of the bases in
the third position of redundant codons. This is done by artificially
synthesizing the DNA sequence of the whole engineered gene. For a whole gene,
individual segments of DNA that overlap are synthesized and then assembled as
discussed in Previews Pages. Genes that have been codon-optimized for new host
organisms may show a 10-fold increase in the level of protein produced, due to
more rapid elongation of the polypeptide chain by the ribosome. This approach
has been used successfully for several of the insect toxins, originally from Bacillus thuringiensis, which have been
cloned into transgenic plants.
Another less labor-intensive
approach to the codon issue is to supply the rare tRNAs. The rarest codons are
not used because the corresponding tRNAs are in short supply. For example, the
rarest codons in E. coli are AGG and
AGA, which both encode arginine and only occur at frequencies of about 0.14%
and 0.21%, respectively. The argU
gene encodes the tRNA that reads both the AGA and AGG codons. If this gene is
supplied on a plasmid, the host E. coli
no longer has a tRNA shortage, and the recombinant proteins can be made.
Plasmids carrying all the genes for rare codon tRNAs are available
commercially. These plasmids have a p15A origin of replication, which makes
them compatible with most expression vectors as these usually have a ColE1
origin of replication.
Related Topics