Chapter: Environmental Biotechnology: Genetic Manipulation
Transgenic Plants
Transgenic Plants
Currently, genetic engineering in agribiotechnology is focusing on genetic mod-ifications to improve crop plants with respect to quality, nutritional value, and resistance to damage by pests and diseases. Other avenues under investigation aim to increase tolerance to extreme environmental conditions, to make plants better suited for their role in pollutant assimilation, degradation or dispersion by phytoremediation, or to modify plants to produce materials which lead to the reduction of environmental pollution. Crop quality improvements such as the control of fruit ripening (Grierson and Schuch 1993), an example of which is the oft quoted, Flavr-Savr tomato, and the production of cereals with improved nutritional value, are not addressed here because, although of great interest to the food industry, are of more peripheral relevance to environmental biotechnology. Many of the transgenic plants, examples of which are given later, have been produced using the Ti plasmid transfer system of Agrobacterium tume-faciensand often, together with the 35S CaMV promoter.
Transformation of plants
There are two practical problems associated with genetic engineering of plants which make them more difficult to manipulate than bacteria. Firstly they have rigid cell walls and secondly they lack the plasmids which simplify so much of genetic engineering in prokaryotes. The first problem is overcome by the use of specialised techniques for transformation, and the second by performing all the manipulations in bacteria and then transferring the final product into the plant. The DNA construct contains regions of DNA which are complementary to the plant DNA to enable the inserted piece to recombine into the plant genome.
The most popular method of transforming plants is by the Ti plasmid but there are at least two other methods also in use. The first is a direct method where DNA is affixed to microscopic bullets which are fired directly into plant tissue. An example of this technology is the introduction into sugarcane, of genes able to inactivate toxins produced by the bacterium, Xanthomonas albilineans, causing leaf scald disease (Zhang, Xu and Birch 1999). This method of biolistic bombardment, may increase in popularity in line with improvements to plastid transformation. It is now possible to produce fertile transgenics expressing foreign proteins in their edible fruit (Ruf et al. 2001).
The second is by protoplast fusion which is a process whereby the plant cell wall is removed leaving the cell surrounded only by the much more fragile membrane. This is made permeable to small fragments of DNA and then the cells allowed to recover and grow into plants. These methods can be unsuccessful due to difficulties in recovery of the cells from the rather traumatic treatments and also because the DNA introduced, has a tendency to be inserted randomly into the genome, rather than at a defined site. However, both methods enjoy the advantage that DNA enters the cell exactly as constructed and has not passed through an intermediate vector giving the opportunity for gene rearrangement.
Transformation by the Ti plasmid of Agrobacterium tumefaciens, shown dia-grammatically in Figure 9.4, suffers from few disadvantages other than the limi-tation that it does not readily infect some cereal crops. This potential problem has been addressed by attempting to increase its host range (Godwin, Fordlloyd and Newbury 1992) which has met with success, leading to improved transformation procedures (Le et al. 2001). In essence, the wild-type plasmid contains genes which causes the transfer of a piece of DNA, ‘T-DNA’, into a plant cell. This piece is bordered by sequences of 24 base pairs in length which are repeats of each other. This structure is fairly common in DNA and is described as direct repeat. The T-DNA comprises genes which cause crown gall disease. These
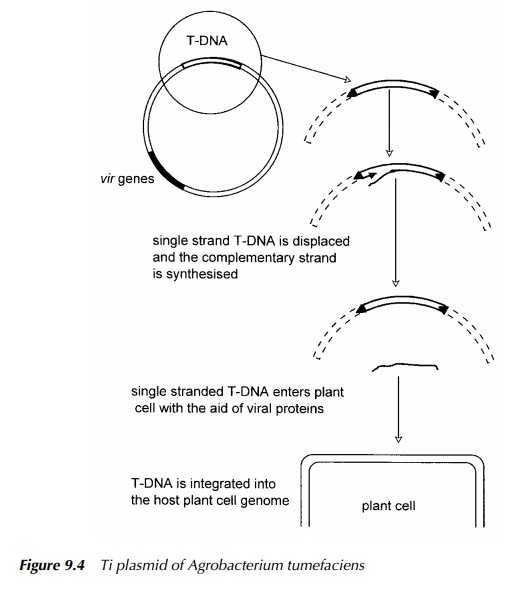
genes may be cut out and replaced by DNA containing the gene of choice to be introduced into the plant. There are many additional elements which may be included in the construct. For example, if the aim is to express the gene, it is pre-ceded by a strong promoter, most commonly the ‘35S’ promoter of Cauliflower Mosaic Virus (CaMV).
In addition to the above, it is important to know if the ‘foreign’ gene is being expressed and so frequently a ‘reporter’ gene described in the section above is also included located close to the gene of interest. Recombination is not 100% efficient, and so a method of selection is required such that only plants con-taining the novel DNA grow. This is frequently a gene coding for resistance to weedkiller or antibiotic. On the grounds of size, this is usually introduced more successfully on a second Ti plasmid during a co-infection with Agrobac-terium carrying the plasmid containing the gene of interest. The experimentcan become somewhat complicated at this stage, as other selector genes are introduced into the plasmids to ensure that growth is only possible if all the desired elements are present in the plant cell. This can involve infection with two or three cultures of Agrobacterium each containing its own engineered Ti plasmid. A very detailed description of the Ti plasmid is published elsewhere (Hughes 1996).
Related Topics