Chapter: Environmental Biotechnology: Genetic Manipulation
Symbiotic nitrogen fixation - Biotechnology Integrated Agricultural Applications
Symbiotic nitrogen fixation
The classic example of plant growth stimulation by plant/microbe
symbiosis is nitrogen fixation by Rhizobium
bacteria within plant root tissue. A full expo-sition of nitrogen fixation may
be found in many classical textbooks and sowill be described here only in
outline. The process of root nodule formation is shown diagrammatically in
Figure 10.5. In total, there is a limited number of organisms able to fix
nitrogen, all of which are prokaryotes. All living organisms are dependent
ultimately on such organisms, due to a universal and essential requirement for
nitrogen, normally in the form of nitrate, ammonia or ammo-nium ion or as amino
acids from which the amino group may be transferred as required. Nitrogen is
fixed by reduction to ammonia either by free living organ-isms or by plant
symbiots. In both cases, it is essential to have an oxygen free environment as
the enzymes involved in the process are irreversibly inactivated by the
presence of oxygen. Of the free living organisms able to fix nitrogen, some achieve
this naturally while others have to create such an environment. Clostridium and
Klebsiella achieve this since they are both anaerobes and so are already
adapted to life in an oxygen-free atmosphere, while Cyanobacteria and Azobacter
have developed means of creating one for themselves. Azobac-ter does this by
having a very high oxygen consumption rate thus effectively creating an
oxygen-free environment. Other nitrogen-fixing bacteria include the filamentous
bacteria such as the Corynebacterium species, and photosynthetic bacteria
referred to elsewhere, such as Rhodospirillum. The latter makes use of
photosynthesis to provide the energy for these reactions and so is presented
with the problem of removing the oxygen produced during photosynthesis away from
nitrogen fixation reactions. Although these free living organisms have a vital
role to play in their particular niches, approximately 10 times more nitrogen
is fixed by plant symbiots. Presumably this is because the plant is better able
to provide the necessary levels of ATP to meet the high energy demands of the
process than are free living bacteria. In addition, the plant supplies the
endophytes with dicarboxylic acids, such as malate and succinate and other
nutrients, like iron, sulphur and molybdenum which is a component of the
nitrogen-fixing enzymes. It also provides its nitrogen-fixing symbiots with an
oxygen-free environment, as described later in this discussion. The analogous
chemical process has an enormous energy requirement to achieve the necessary
high temperature, in the region of 500 ◦ C, and a pressure in excess of 200
atmospheres. This is part of the reason why manufacture of fertilisers for
agricultural purposes, which is in effect the industrial equivalent of the
biological process, is a drain on natural resources. Together with the
unwelcome leaching of surplus fertiliser into water-ways, causing algal blooms
among other disturbances, this makes the widespread application of fertiliser
recognised as a potential source of environmental dam-age. It is understandable
to see a drive towards the engineering of plants both to increase the
efficiency of nitrogen fixation, which is estimated at being 80% efficient, and
to extend the range of varieties, and especially crop species, which have this
capability. It is also worth noting that unlike superficially applied
fer-tilisers which may exceed locally the nitrogen requirement and so leach
into the surrounding waterways, nitrogen fixation by bacteria occurs only in
response to local need and so is very unlikely to be a source of pollution.
The following brief
description of the required interactions between plants and microbes, the
example used here being Rhizobium,
should serve to illustrate why such a goal is very difficult to achieve.
Firstly, the plant is invaded by a member of the Rhizobium family of free living soil bacteria. There is a specific
relationship between plant and bacterium such that only plants susceptible to
that particular member of the group may be infected. Genes involved in the
infection process and nodule formation called nod genes are coded for by the bacterium. The nod genes are activated by a mixture of flavenoids released by the
plant into the region around the roots and thus the plant signals to the
bacterium its receptiveness to be infected.
After infection through the
root hairs, the multiplying bacteria find their way into the cells of the inner
root cortex. They are drawn into the cell by endo-cytosis shown in Figure 10.5
and so are present within the cell bounded by
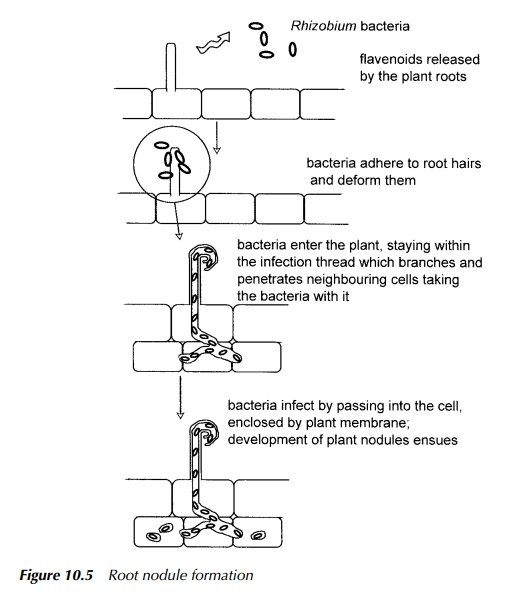
plant cell membrane. This structure then develops into nodules
containing the nitrogen-fixing bacteria. Several changes to the plant then
ensue including the synthesis of proteins associated with the nodule, the most
abundant of which is leghaemoglobin, which may reach levels of up to 30% of the
total nodule protein. The genes coding for this protein are partly bacterial
and partly plant in origin and so exemplify the close symbiosis between the two
organisms. The expres-sion of leghaemoglobin is essential for nitrogen
fixation, since it is responsible for the control of oxygen levels. The enzymes
for fixation are coded for by a plasmid of Rhizobium
and are referred to as the nif gene
cluster. There are two components each comprising a number of genes. One
component is nitrogenasereductase the
function of which is to assimilate the reducing power used by thesecond
component, nitrogenase, to reduce
nitrogen to ammonium ion. Expression of the nif
genes is highly regulated in all organisms studied to date. In addition to the nif and the nod genes, Rhizobium also
carries additional genes which are involved in the fixation of nitrogen called
the fix genes. Once ammonia, or the
ammonium ion, has been formed it may be transferred to the amino acid,
glutamate, to produce glutamine. This transfer is frequently invoked throughout
metabolism. This is not the only route for the assimilation of the amino group
into metabolic pathways; the synthesis of the purine derivative, allantoin or
allantoic acid, being a less heavily used alternative. These pathways are
described in Chap-ter 2. Nitrogen fixation involving Rhizobium has been the focus of the preceding discussion, but the
family of aerobic, Gram positive bacteria, Actinomycetes, is another group
carrying out the same function using a similar mechanism. These may form a
network of aerial or substrate hyphae resembling a structure com-monly
associated with that of fungi. These bacteria may form a close association,
called actinorhizae, with the roots of a number of plants which tend to be
woody, or shrub-like in nature.
Related Topics