Chapter: Genetics and Molecular Biology: DNA Synthesis
The DNA Elongation Rate - Physiological Aspects
The DNA Elongation Rate
It seems reasonable that half the single bacterial
chromosome should be replicated by a single replication region traversing the
chromosome in about one doubling time. If such a replication region does not
contain multiple points of DNA elongation, then the speed of DNA chain
elon-gation must be on the order of 1,000 nucleotides per second to replicate
the chromosome’s 3 × 106 bases in a typical doubling time
for rapidly growing bacteria of 30 minutes. Measuring such a rate is very
difficult, but fortunately it is possible to reduce its value by about a factor
of 5 by growing the cells at 20° instead of 37°, the temperature of most rapid
growth.
If radioactive DNA precursors are added to the cell
growth medium and growth is stopped soon thereafter, the total amount of
radioactivity, T, incorporated into
DNA equals the product of four factors: a constantrelated to the specific
activity of the label, the number of growing chains, the elongation rate of a
chain, and the time of radioactive labeling T
=c×N× R × t. Similarly, the total amount of radioactivity
incorpo-rated into the ends of elongating chains, E, equals the product of two
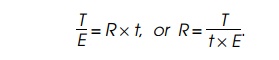
Determination of the T radioactivity is straightforward, but determi-nation of the end
radioactivity is not so obvious. Furthermore, careful experimentation would be
required so that losses from either T
or E samples do not introduce errors.
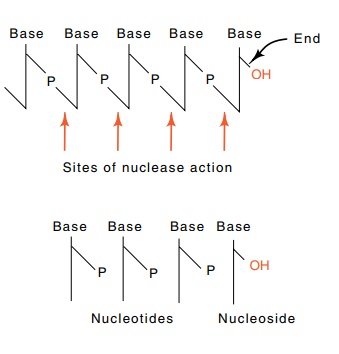
These problems can be solved by digesting the DNA extracted from the labeled
cells with a nuclease that cleaves so as to leave a phosphate on the 3’
position of the deoxyribose. After the digestion, the terminal deoxyribose from
the elongating chain lacks a phosphate, whereas the internal deoxyriboses all
possess phos-phate groups (Fig. 3.15).
Hence separation and quantitation of the radioactive nucleosides and
nucleotides in a single sample prepared from cells following a short
administration of the four radioactive DNA precursors yield the desired T and E values.
If the elongation rate is several hundred bases per
second, then one second of synthesis will label several hundred bases, and a
separation of nucleosides and nucleotides will then need to be better than one
part in several hundred. Also, it is difficult to add label suddenly and then
to stop the cells’ DNA synthesis quickly. Finally, the specific activity of
intracellular nucleoside triphosphate pools does not immediately jump to the
same specific activity as the label added to the medium. Fortu-nately the
effect of a changing specific activity can be simply accounted for by taking a
series of samples for analysis at different times after the addition of
radioactive label.
The sample from the first point that could be taken from the cells after the addition of radioactive label possessed little radioactivity. It had counts of 17-20 cpm in ends and 2-20 × 105 cpm in total DNA. Samples taken later possessed greater amounts of radioactivity. This experiment yielded elongation rates of 140-250 bases/sec in cells with a doubling time of 150 minutes. This corresponds to a rate of 400 to 800 bases/sec in cells growing at 37° with a doubling time of 45 minutes. Therefore, only about two elongation points exist per replication region. The effects on this type of measurement of discontinuous DNA replica-tion on the lagging strand are left as a problem for the reader.
The primary conclusion that should be drawn from
the direct meas-urement of the DNA elongation rate is that a small number of
enzyme molecules are involved. The cell does not utilize an extensive factory
with many active DNA polymerase molecules in a growing region. What happens
when cells grow at 37° but at different growth rates due to the presence of
different growth media?
Related Topics