Chapter: Genetics and Molecular Biology: DNA Synthesis
Bidirectional Replication from E. coli Origins - Physiological Aspects
Bidirectional Replication from E. coli Origins
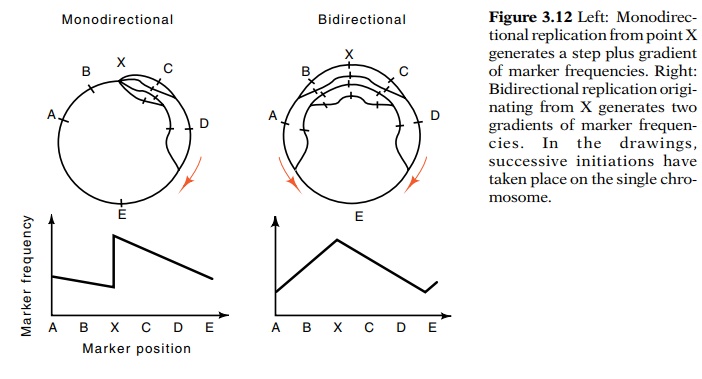
The Masters and Broda experiment was designed to locate the origin of replication on the genetic map. It is primarily a genetic experiment, but it utilizes the fact discussed that an exponentially growing population contains more young individuals than old individu-als. Chromosomes can be considered in the same way. Correspondingly, a population of growing and dividing chromosomes contains more members just beginning replication than members just finishing repli - cation. The Cairns autoradiograph experiments show that the chromosome is replicated sequentially.
A population of
cells or chromosomes that is growing exponentially will contain more copies of
genes located near the origin of replication than genes located at the terminus
of replication. This same idea was used to locate the replication origin and
demonstrate bidirectional replication of the SV40 animal virus.
Determining whether the bacterial chromosome is
replicated in one direction from a unique origin or in both directions from a
unique origin becomes a question of counting gene copies. If we could count the
number of copies of genes A, B, C, D, and E, we could determine whether the
cell uses monodirectional or bidirectional DNA replication initiating from
point X (Fig. 3.12).
Several methods exist for counting the relative numbers
of copies of different genes or chromosome regions. Here we shall consider a
bio-logical method for performing such counting that utilizes the phage P1.
This method depends upon the fact that after P1 infection, a cell synthesizes
about 100 new P1 particles. Most of these package their own DNA. A few phage
particles package E. coli DNA
instead. If a P1 lysate that was prepared on one type of cells is then used to
infect a second culture of cells, most of the infected cells will proceed to
make new phage P1. Those few cells that are infected with a P1 coat containing E. coli
DNA from the first cells may be able to recombine that particularstretch of
E. coli DNA into their chromosomes. By this means they can replace
stretches of chromosomal DNA with chromosomal DNA brought into them by the
phage particles (Fig. 3.13). This process is termed transduction.
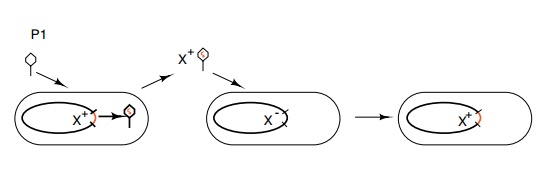
Figure 3.13 Transduction of genetic markers by phage P1 infecting an X+celland carrying the X+ marker into an X- cell.
The numbers of these defective phage particles
carrying different genes from the infected cells is related to the numbers of
copies of these genes present at the time of phage infection. Transduced cells
can be made to reveal themselves as colonies, thereby permitting their simple
and accurate quantitation. Consequently the use of phage P1 enabled counting of
the relative numbers of copies within growing cells of various genes located
around the chromosome. The results together with the known genetic map
indicated that E. coli replicates its chro-mosome
bidirectionally and determined the genetic location of the replication origin.
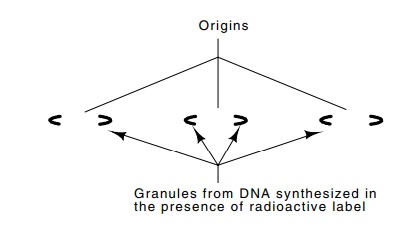
Figure
3.14 Schematic of an electron
micrograph of an autoradiograph pre-pared from mouse DNA extracted after 30
seconds of labeling with radioactive thymidine. DNA is replicated
bidirectionally from the multiple origins.
On the opposite side of the chromosome from the
origin lies a terminus region. A protein called Tus binds to the terminus. It
blocks elongation by inactivating the oncoming helicase of the replication
fork.
Autoradiographic experiments have also suggested
that mammalian DNA also replicates bidirectionally from origins (Fig. 3.14).
Multiple replication forks or “eyes” could be shown to derive from a single DNA
duplex. The replication trails indicated that two forks originated from a
single origin. Furthermore, the rate of elongation of labeled strands showed
that the synthesis rate of mammalian DNA is about 200 nucleo-tides per second.
Related Topics