Chapter: Genetics and Molecular Biology: Nucleic Acid and Chromosome Structure
Southern Transfers to Locate Nucleosomes on Genes
Southern Transfers to Locate Nucleosomes on
Genes
Southern transfers are a versatile technique of
molecular genetics that combines electrophoresis and DNA-DNA or RNA-DNA
hybridization
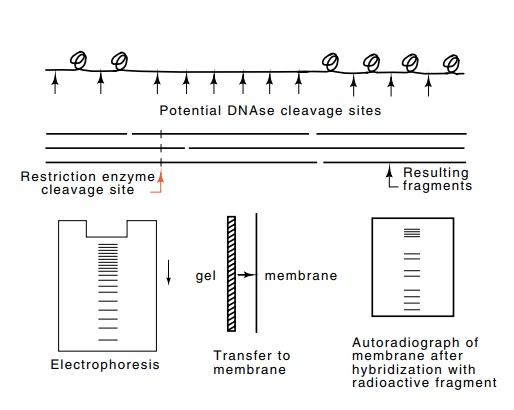
Figure
2.15 Use of Southern transfer
technology to determine the locations ofnuclease hypersensitive and
hyposensitive sites in the vicinity of a gene. Gently extracted DNA is lightly
digested with DNAse, cleaved with a restriction enzyme at specific sites,
denatured, separated by electrophoresis, transferred to a membrane, and then
hybridized to a short radioactive oligonucleotide that hybridizes near the gene
in question. Finally, an autoradiograph is made of the membrane.
The application of Southern transfers
to various questions will be mentioned a number of this throughout the book.
The power of the transfer and hybridization technology as applied to examining
nucleosome positioning is that it permits determination of the suscep-tibility
to DNAse cleavage in the region of whatever gene we are interested in. This can
be done in the presence of DNA from thousands of other genes. Here we shall
consider the application of this technology to the problems of ascertaining
whether nucleosomes near a specific gene occupy fixed positions and whether
nucleosomes cover regulatory sequences just ahead of genes. The approach has
shown that in front of many genes are areas apparently not occupied by
nucleosomes. This leaves these regions hypersensitive to hydrolysis by
nucleases added to gently lysed nuclei. The nuclease sensitivity within the
genes is much less due to the presence of nucleosomes. Often such nucleosomes
tend to occupy specific positions.
The DNA for nucleosome position measurements is
gently extracted from nuclei and lightly treated with a nuclease like DNAse I
to generate about one nick per one thousand base pairs. Different molecules
will be nicked in different places, but very few molecules will be nicked in
areas covered by nucleosomes. After the digestion, protein is removed by
extraction with phenol and all the DNA molecules are digested by anenzyme that
cleaves DNA at specific sequences. These enzymes are known as restriction
enzymes and will be discussed more fully later.
Suppose that such a cleavage site lies several
hundred base pairs in front of the gene we are considering. After the cleavage
steps, the DNA fragments are denatured and the single-stranded fragments are
sepa-rated according to size by electrophoresis. After the electrophoresis, the
fragments are transferred to a sheet of nylon membrane. The transfer to the
membrane preserves the pattern of size-separated fragments. The membrane can
then be incubated in a solution containing radioactive oligonucleotide
possessing a sequence complementary to sequence from the gene of interest near
to the cleavage site. The oligonucleotide will hybridize to just those DNA
fragments possessing this complemen-tary sequence. Hence the membrane will be
radioactive in the areas containing the fragments. In any area of the DNA that
was protected from DNAse I nicking by the presence of a nucleosome, no
cleavages will occur. Hence, there will be no fragments of the size extending
from the position of the restriction enzyme cleavage site to the area occupied
by the nucleosome. Conversely, in areas readily cleaved by the nuclease, many different
molecules will be cleaved, and therefore many DNA fragments will exist of a
length equal to the distance from the restriction cleavage site to the
nuclease-sensitive, nucleosome free, region.
Nucleosome protection experiments show that several
hundred nu-cleotides in regions ahead of genes in which regulatory proteins are
expected to bind frequently are devoid of nucleosomes. Two factors are
responsible. First, regulatory proteins can bind to these regions and prevent
nucleosomes from binding there. Another reason is natural bending of the DNA.
As discussed earlier, DNA is not straight, and most DNA possesses minor bends.
Such bends greatly facilitate the wrapping of DNA around the histones in the
formation of a nucleosome. Thus, bends in the DNA can position a nucleosome,
and this in turn partially positions its neighbors, generating a region of
phased nucleosomes. Such phasing can leave gaps where they are necessary for
the binding of regulatory proteins.
Related Topics