Chapter: Genetics and Molecular Biology: Nucleic Acid and Chromosome Structure
Altering Linking Number
Altering Linking Number
`Cells have enzymes that alter the linking numbers
of covalently closed DNA molecules. Wang, who found the first such protein,
called it omega, but now it is often called DNA topoisomerase I. Astoundingly,
this enzyme removes negative superhelical turns, one at a time, without
hydrolyzing ATP or any other energy-rich small molecule. It can act on
positively supercoiled DNA, but only if a special trick is used to generate a
single-stranded stretch for the enzyme to bind to. No nicks are left in
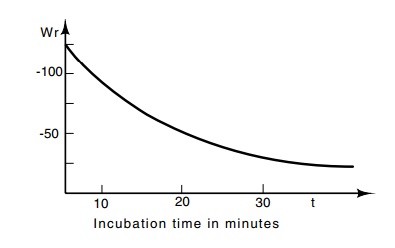
Figure
2.13 Reduction in the number of
negative superhelical turns containedin lambda phage DNA as a function of the
time of incubation with DNA topoisomerase I.
the DNA after omega has acted. Purified, covalently
closed lambda phage DNA extracted from lambda-infected cells is found to
possess about 120 negative superhelical turns. Incubation of omega with this
DNA increases the linking number until only about 20 negative super-helical
turns remain (Fig. 2.13). The enzyme appears to remove twists, and hence
superhelical turns, in a controlled way. It binds to the DNA and then
interrupts the phosphodiester backbone of one strand by a phosphotransfer
reaction that makes a high-energy phosphate bond to the enzyme. In this state
the enzyme removes one twist and then re-forms the phosphodiester bond in the
DNA.
Additional enzymes capable of altering the linking
number of DNA have also been discovered. The topoisomerase from eukaryotic
cells can apparently bind double-stranded DNA and will remove positive super-helical
turns as well as negative. Cells possess another remarkable enzyme. This
enzyme, DNA gyrase or DNA topoisomerase II, adds negative superhelical turns;
as expected, energy from the hydrolysis of ATP is required for this reaction.
Studies of the activity of topoisomerases I and II,
omega and gyrase, have shown that these enzymes can best be thought of as
functioning by strand passage mechanisms. Where two DNA duplexes cross, gyrase,
with the consumption of an ATP molecule, can permit the duplex lying below to
be cut, to pass the first, and to be rejoined. Topoisomerase I
can proceed by the same sort of pathway, but it
passes one strand of a duplex through the other strand, having first denatured
a region where
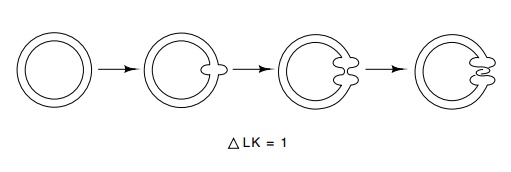
Figure
2.14 The strand inversion pathway as
applied by DNA topoisomeraseI in which a region of DNA is melted and one strand
is broken, passed by the other, and rejoined.
strand crossing is to occur (Fig. 2.14). These are
known as sign-inversion pathways because they change the sign of the linking
number that is contributed by a point of crossing of the two DNA strands.
The sign-inversion pathway has a particularly
useful property: it permits DNA duplexes to interpenetrate one another. Gyrase
is an enzyme that can untangle knots in DNA! Undoubtedly this property is of
great value to the cell as the DNA is compressed into such a small volume that
tangles seem inevitable. In general, enzymes that cut and rejoin DNA are
topoisomerases.
Related Topics