Chapter: Genetics and Molecular Biology: Nucleic Acid and Chromosome Structure
Grooves in DNA and Helical Forms of DNA
Grooves in DNA and Helical Forms of DNA
Watson and Crick deduced the basic structure of DNA
by using three pieces of information: X-ray diffraction data, the structures of
the bases, and Chargaff’s findings that, in most DNA samples, the mole fraction
of guanine equals that of cytosine, as well as the mole fraction of adenine
equals that of thymine. The Watson-Crick structure is a pair of oppositely
oriented, antiparallel, DNA strands that wind around one another in a
right-handed helix. That is, the strands wrap clockwise moving down the axis
away from an observer. Base pairs A-T and G-C lie on the interior of the helix
and the phosphate groups on the outside.
In semicrystalline fibers of native DNA at one
moisture content, as well as in some crystals of chemically synthesized DNA,
the helix repeat is 10 base pairs per turn. X-ray fiber diffraction studies of
DNA in different salts and at different humidities yield forms in which the
repeats vary from 9 1⁄3 base
pairs per turn to 11 base pairs per turn. Crystallographers have named the different
forms A, B, and C (Table 2.1).
More recent diffraction studies of crystals of
short oligonucleotides of specific sequence have revealed substantial base to
base variation in the twist from one base to the next. Therefore, it is not at
all clear
The A, B, and C forms of DNA are all helical. That
is, as the units of phosphate-deoxyribose-base:base-deoxyribose-phosphate along
the DNA are stacked, each succeeding base pair unit is rotated with respect to
the preceding unit. The path of the phosphates along the periphery of the
resulting structure is helical and defines the surface of a circular
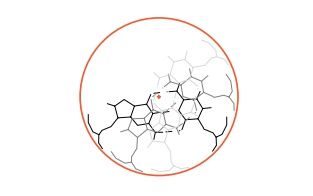
cylinder just enclosing the DNA. The base pair unit
is not circular, however, and from two directions it does not extend all the
way to the enclosing cylinder. Thus a base pair possesses two indentations.
Be-cause the next base pair along the DNA helix is rotated with respect to the
preceding base, its indentations are also rotated. Thus, moving along the DNA
from base to base, the indentations wind around the cylinder and form grooves.
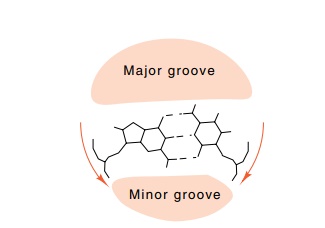
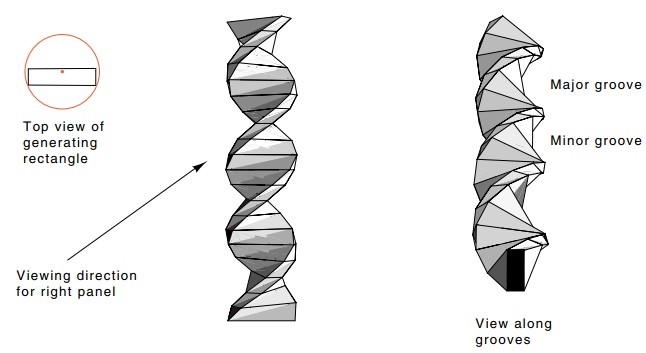
Fig. 2.3 shows a helix generated from a rectangle
approximating the base pair unit of B DNA. Note that the rectangle is offset
from the helix axis. As a result of this offset, the two grooves generated in
the helix are of different depths and slightly different widths. The actual
widths of the two grooves can be seen more clearly from the side view of the
three dimensional helix in which the viewpoint is placed so that you are
looking directly along the upper pair of grooves.
Figure 2.3 Generation of a helix by the stacking of rectangles. Each rectangleis rotated 34° clockwise with respect to the one below. Left, the viewpoint and the generating rectangle; right, a view along the major and minor grooves showing their depths and shapes.
In actual DNA, the deoxyribose-phosphate units are
not aligned parallel to the base pairs as they were represented in the
rectangle approximation above. The units are both oriented toward one of the
grooves. This narrows one of the grooves on the helical DNA molecule and widens
the other. The two grooves are therefore called the minor and major grooves of
the DNA. Thus, the displacement of the base pair from the helix axis primarily
affects the relative depth of the two grooves, and the twisted position of the
phosphates relative to the bases primarily affects the widths of the grooves.
The A-form of DNA is particularly interesting
because the base pairs are displaced so far from the helix axis that the major
groove becomes very deep and narrow and the minor groove is barely an
indentation. Helical RNA most often assumes conformations close to the A-form.
Additional factors such as twist and tilt of the base pairs which are
present in the A and C-forms of DNA, but largely
absent in the B-form, have lesser effects on the width and depth of the
grooves. Unexpectedly, even a left-helical form can form under special
conditions, but thus far this Z-form DNA has not been found to play any
significant biological role.
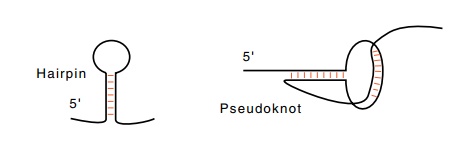
Helical structures can also form from a single
strand of RNA or DNA if it folds back upon itself. The two most common structures
are hairpins and pseudoknots. In a pseudoknot, bases in the loop form
additional base pairs with nucleotides beyond the hairpin region.
Related Topics