Chapter: Biotechnology Applying the Genetic Revolution: Aging and Apoptosis
Sirtuins, Histone Acetylation, and Life Span in Yeast
SIRTUINS,
HISTONE ACETYLATION, AND LIFE SPAN IN YEAST
Despite being a single-celled eukaryote,
yeast undergoes aging of two different types, replicative aging and
chronological aging. Replicative aging applies to dividing cells and is
measured by the number of daughter cells one mother can produce. Chronological
aging is induced by nutrient deprivation. The length of time the cell can live
without dividing determines its chronological age.
Aging in yeast is affected by the Sir2
protein, a member of the sirtuin family of histone deacetylases. (Remember that
eukaryotic gene expression involves remodeling of the nucleosomes, which in
turn is affected by the level of histone acetylation.) Yeast cells that lack
Sir2 have a longer chronological life span. They have a decreased rate of DNA
mutation and increased resistance to stressful conditions such as heat shock
and oxidative stress. The mammalian homolog to Sir2, SirT1, controls glucose
and fat metabolism. Mammalian cells lacking the sirt1 gene survive
longer in culture, and transgenic mice lacking this gene have lower body fat
and live longer. Sir2 is thus implicated in the caloric restriction pathway
discussed earlier.
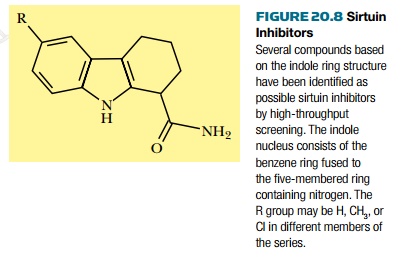
Inhibitors of sirtuins are being
developed as possible anti-aging drugs. Indeed, one of the biotech companies
involved is named Elixir Pharmaceuticals. They have developed a series of
modified indoles as specific and potent sirtuin inhibitors ( Fig. 20.8 ).
Sirtuins act by transferring the acetyl group from the histone to NAD, giving O
-acetyl-ADP-ribose plus nicotinamide. The indolebased sirtuin inhibitors
bind after the release of nicotinamide and prevent the release of the histone
and the O -acetyl-ADP-ribose.
Related Topics