Chapter: Biochemistry: Biosynthesis of Nucleic Acids: Replication
Replication of DNA
Replication of DNA
Naturally
occurring DNA exists in many forms. Single- and double-stranded DNAs are known,
and both can exist in linear and circular forms. As a result, it is difficult
to generalize about all possible cases of DNA replication. Since many DNAs are
double-stranded, we can present some general features of the replication of
double-stranded DNA, features that apply both to linear and to circular DNA.
Most of the details of the process that we shall discuss here were first
investigated in prokaryotes, particularly in the bacterium Escherichia coli. We shall use information obtained by experiments
on this organism for most of our discussion of the topic.
The
process by which one double-helical DNA molecule is duplicated to produce two
such double-stranded molecules is complex. The very complexity allows for a
high degree of fine-tuning which, in turn, ensures considerable fidelity in
replication. The cell faces three important challenges in carrying out the
necessary steps. The first challenge is separating
the two DNA strands. The two strands of DNA are wound around each other in
such a way that they must be unwound if they are to be separated. In addition
to achieving continuous unwinding of the double helix, the cell also must
protect the unwound portions of DNA from the action of nucleases that preferentially attack single-stranded DNA. The
second task involves synthesizing of DNA
from the 5' to the 3' end. Two antiparallel strands must be synthesized in
the same direction on antiparallel templates. In other words, the template has
one 5' - > 3' strand and one 3' - > 5' strand, as does the newly
synthesized DNA. The third task is guarding
againsterrors in replication, ensuring that the correct base is added to
the growing poly-nucleotide chain.
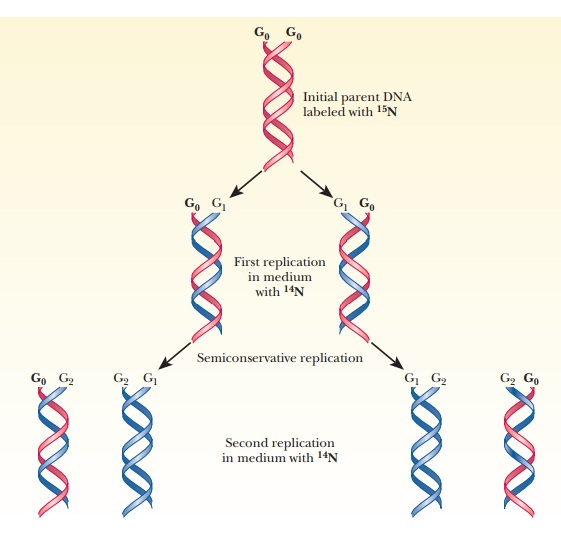
Semiconservative Replication
DNA
replication involves separation of the two original strands and production of
two new strands with the original strands as templates. Each new DNA molecule
contains one strand from the original DNA and one newly synthesized strand.
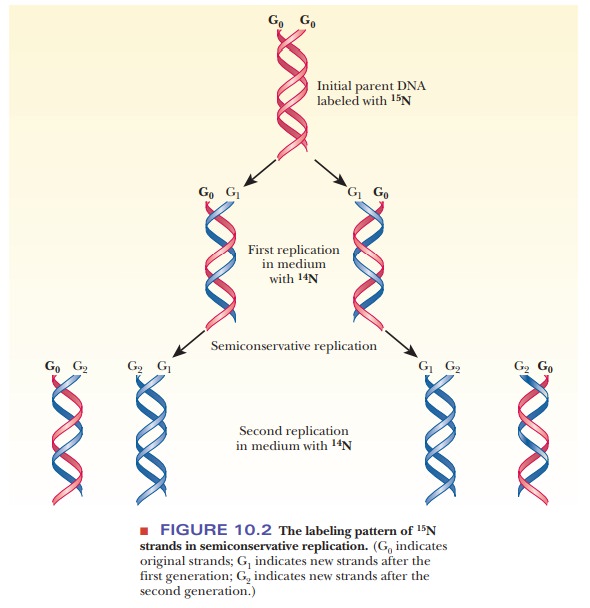
This situation is called semiconservative
replication (Figure 10.2). The details of the process differ in prokaryotes
and eukaryotes, but the semiconservative nature of replication is observed in
all organisms.
How did scientists figure out that replication is semiconservative?
Semiconservative replication of DNA was established unequivocally in the late 1950s by experiments performed by Matthew Meselson and Franklin Stahl. E. coli bacteria were grown with15NH4Cl as the sole nitrogen source,15N beinga heavy isotope of nitrogen. (The usual isotope of nitrogen is 14N.) In such a medium, all newly formed nitrogen compounds, including purine and pyrimidine nucleobases, become labeled with 15N.
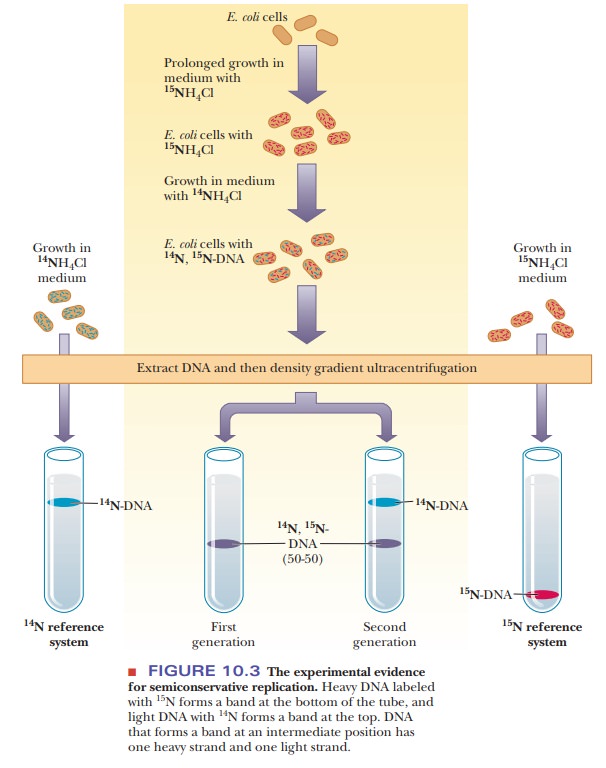
The 15N-labeled
DNA has a higher density than unlabeled DNA, which contains the usual isotope, 14N. In
this experiment, the 15N-labeled cells were then
transferred to a medium that contained only 14N. The
cells continued to grow in the new medium. With every new generation of growth,
a sample of DNA was extracted and analyzed by the technique of density-gradient centrifugation (Figure
10.3). This technique depends on the fact that heavy 15N DNA
(DNA that contains 15N alone) forms a band at the
bottom of the tube; light 14N DNA (containing 14N alone)
appears at the top of the tube. DNA containing a 50–50 mixture of 14N and 15N
appears at a position halfway between the two bands. In the actual experiment,
this 50–50 hybrid DNA was observed after one generation, a result to be
expected with semiconservative replication. After two generations in the
lighter medium, half of the DNA in the cells should be the 50–50 hybrid and
half should be the lighter 14N DNA. This prediction of the
kind and amount of DNA that should be observed was confirmed by the experiment.
In which direction does replication go?
During replication, the DNA double helix unwinds at a specific point called the origin of replication (OriC inE. coli). New polynucleotide chains are synthesizedusing each of the exposed strands as a template. Two possibilities exist for the growth of the new strands: synthesis can take place in both directions from the origin of replication, or in one direction only. It has been established that DNA synthesis is bidirectional in most organisms, with the exception of a few viruses and plasmids. (Plasmids are rings of DNA that are found in bacteria and that replicate independently from the regular bacterial genome.). For each origin of replication, there are two points (replication forks) at which new polynucleotide chains are formed.
A “bubble”(also called an “eye”) of newly
synthesized DNA between regions of the original DNA is a manifestation of the
advance of the two replication forks in opposite directions. This feature is
also called a θ
structure because of its resemblance to the lowercase Greek letter theta.
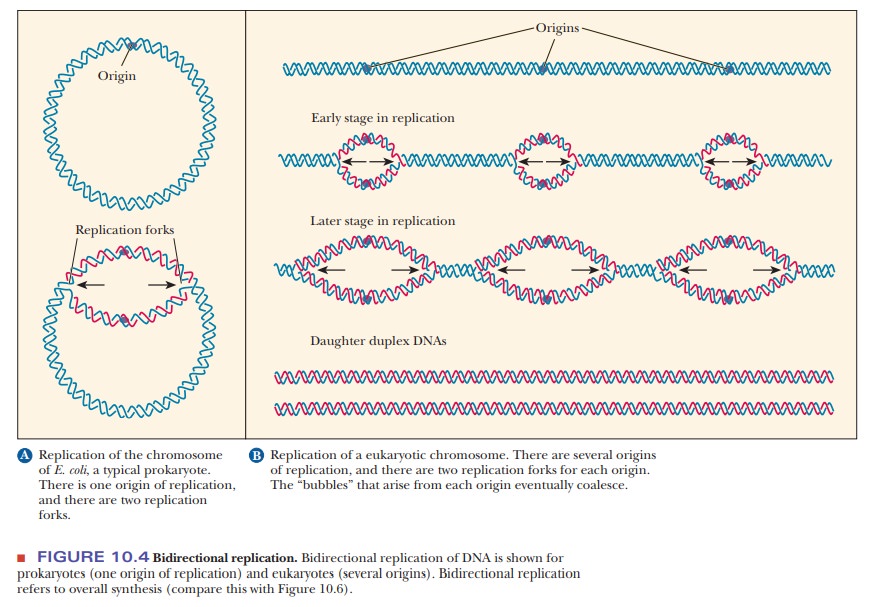
One such
bubble (and one origin of replication) exists in the circular DNA of
prokaryotes (Figure 10.4a). In eukaryotes, several origins of replication, and
thus several bubbles, exist (Figure 10.4b). The bubbles grow larger and
even-tually merge, giving rise to two complete daughter DNAs. This
bidirectional growth of both new polynucleotide chains represents net chain growth. Both new
polynucleotide chains are synthesized in the 5' to 3' direction.
Summary
When a molecule of DNA is
replicated, each of the two strands is used as a template to create a
complementary strand. When a cell divides into two, each of the two cells has
one of the original template strands and one of the new strands. This process
is called semiconservative replication.
When DNA molecules are replicated, the strands are separated at origins of replications. Synthesis occurs in both directions from the origin along replication forks.
Related Topics