Chapter: Biochemistry: Biosynthesis of Nucleic Acids: Replication
DNA Polymerase
DNA Polymerase
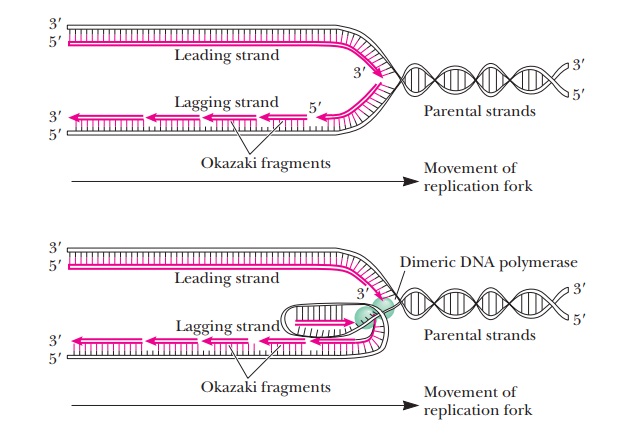
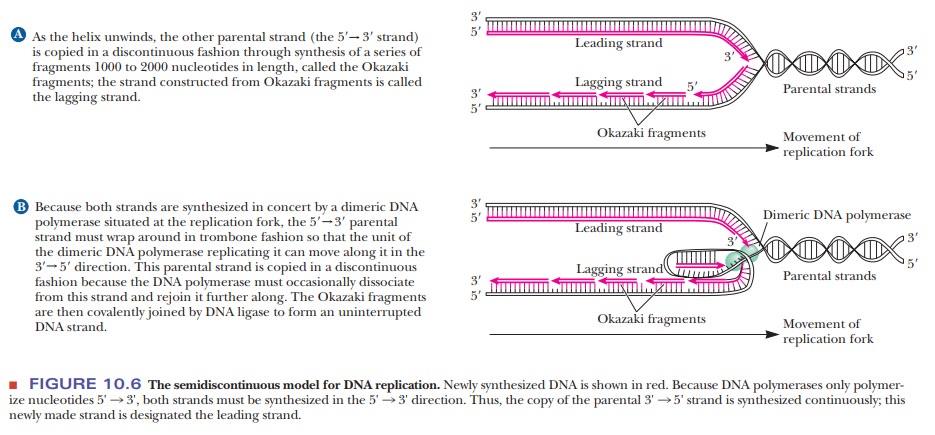
Semidiscontinuous DNA Replication
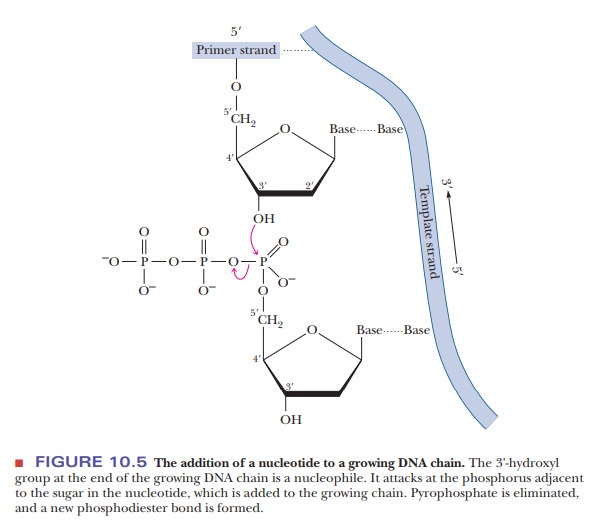
All synthesis of nucleotide chains occurs in the 5' - > 3' direction from the perspective of the chain being synthesized. This is due to the nature of the reaction of DNA synthesis. The last nucleotide added to a growing chain has a 3'-hydroxyl on the sugar. The incoming nucleotide has a 5'-triphosphate on its sugar. The 3'-hydroxyl group at the end of the growing chain is a nucleophile. It attacks the phosphorus adjacent to the sugar in the nucleotide to be added to the growing chain, leading to the elimination of the pyrophosphate and the formation of a new phosphodiester bond (Figure 10.5). We discussed nucleophilic attack by a hydroxyl group at length in the case of serine proteases; here we see another instance of this kind of mechanism. It is helpful to always keep this mechanism in mind. The further in depth we study DNA, the more the directionality of 5' - > 3' can lead to confusion over which strand of DNA we are discussing. If you always remember that all synthesis of nucleotides occurs in the 5' - > 3' direction from the perspective of the growing chain, it will be much easier to understand the processes to come.
This
universal nature of synthesis presents a problem for the cell because as the
DNA synthesis proceeds along a replication fork, the two strands are going in
opposite directions.
How can replication proceed along the DNA if the two strands are going in opposite directions?
The
problem is solved by different modes of polymerization for the two growing
strands. One newly formed strand (the leading strand) is formed continuously
from its 5' end to its 3' end at the replication fork on the exposed 3' to 5'
template strand. The other strand (the lagging strand) is formed
semidiscontinuously in small fragments (typically 1000 to 2000 nucleotides
long), sometimes called Okazaki fragments after the scientist who first studied
them (Figure 10.6). The 5' end of each of these fragments is closer to the
replication fork than the 3' end. The fragments of the lagging strand are then
linked together by an enzyme called DNA
ligase.
DNA Polymerase from E. coli
The first DNA polymerase discovered was found in E. coli.DNA polymerase catalyzes the successive addition of each new nucleotide to the growing chain.
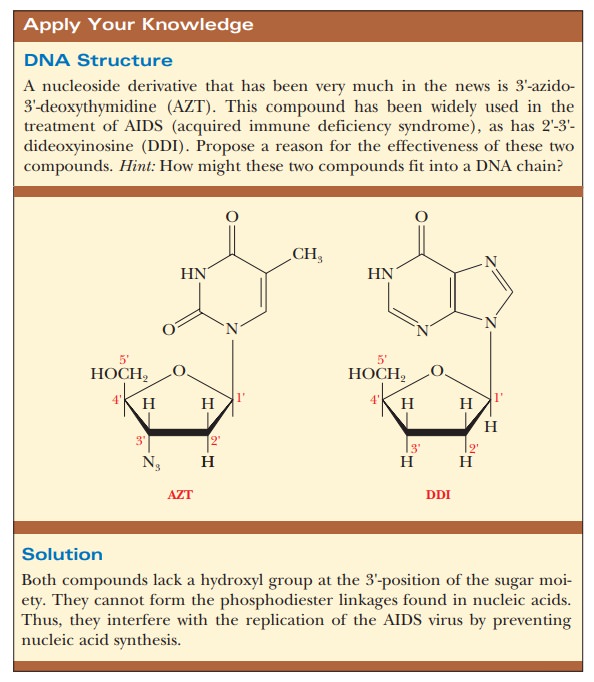
At least
five DNA polymerases are present in E.
coli. Three of them have been studied more extensively, and some of their
properties are listed in Table 10.1. DNA polymerase I (Pol I) was discovered
first, with the subsequent discovery of polymerases II (Pol II) and polymerase
III (Pol III). Polymerase I consists of a single polypeptide chain, but
polymerases II and III are multisubunit proteins that share some common
subunits. Polymerase II is not required for replication; rather, it is strictly
a repair enzyme. Recently, two more polymerases, Pol IV and Pol V, were discovered.
They, too, are repair enzymes, and both are involved in a unique repair
mechanism called the SOS response Two important considerations regarding the
polymerases are the speed of the synthetic reaction (turnover number) and the processivity, which is the number of
nucleotides joined before the enzyme dissociates from the template (Table
10.1).

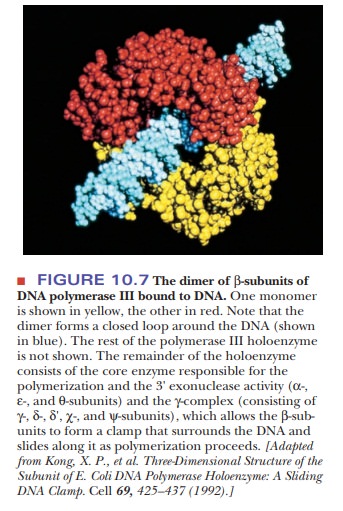
Polymerase III consists of a core enzyme responsible for the polymerization and 3' exonuclease activity-consisting of α-, ε-, and θ-subunits-and a number of other subunits, including a dimer of α-subunits responsible for DNA bind-ing, and the γ-complex-consisting of γ-, δ-, δ', χ-, and ψ-subunits-which allows the β-subunits to form a clamp that surrounds the DNA and slides along it as polymerization proceeds (Figure 10.7). Table 10.2 gives the subunit composi-tion of the DNA polymerase III complex. All of these polymerases add nucleo-tides to a growing polynucleotide chain but have different roles in the overall replication process.
As can be seen in Table 10.1, DNA polymerase III has the highest
turnover number and a huge processivity compared to polymerases I and II.
If DNA
polymerases are added to a single-stranded DNA template with all the
deoxynucleotide triphosphates necessary to make a strand of DNA, no reaction
occurs. It was discovered that DNA polymerases cannot catalyze de novo
synthesis. All three enzymes require the presence of a primer, a short oligonucleotide strand to which the growing
polynucleotide chain is covalently attached in the early stages of replication.
In essence, DNA polymerases must have a nucleotide with a free 3'-hydroxyl
already in place so that they can add the first nucleotide as part of the
growing chain. In natural replication, this primer is RNA.
DNA
polymerase reaction requires all four deoxyribonucleoside triphos-phates-dTTP,
dATP, dGTP, and dCTP. Mg2+ and a DNA template are also
necessary. Because of the requirement for an RNA primer, all four
ribonucleo-side triphosphates-ATP, UTP, GTP, and CTP-are needed as well; they
are incorporated into the primer. The primer (RNA) is hydrogen-bonded to the
template (DNA); the primer provides a stable framework on which the nascent chain
can start to grow. The newly synthesized DNA strand begins to grow by forming a
covalent linkage to the free 3'-hydroxyl group of the primer.
It is
now known that DNA polymerase I has a specialized function in replication-repairing
and “patching” DNA-and that DNA polymerase III is the enzyme primarily
responsible for the polymerization of the newly formed DNA strand. The major
function of DNA polymerases II, IV, and V is as repair enzymes. The exonuclease
activities listed in Table 10.1 are part of the proofreading-and-repair
functions of DNA polymerases, a process by which incorrect nucleotides are
removed from the polynucleotide so that the cor-rect nucleotides can be
incorporated. The 3' - > 5' exonuclease activity, which all three
polymerases possess, is part of the proofreading
function; incorrect nucleotides are removed in the course of replication and
are replaced by the correct ones. Proofreading is done one nucleotide at a
time. The 5' - > 3' exo-nuclease activity clears away short stretches of nucleotides
during repair, usu-ally involving
several nucleotides at a time. This is also how the RNA primers are removed.
The proofreading-and-repair function is less effective in some DNA polymerases.
Summary
To achieve 5' - > 3' synthesis of DNA on two
strands that are antiparallel, DNA polymerase synthesizes one strand
continuously and the other dis-continuously.
The strand synthesized continuously is called the leading strand,
and the one synthesized discontinuously is called the lagging strand.
The pieces of DNA formed discontinuously are called Okazaki
fragments, and they are later joined together by DNA ligase.
The reaction of DNA synthesis involves the
nucleophilic attack of the 3'-hydroxyl of one nucleotide on the γ-phosphate of the incoming nucleo-side
triphosphate.
At least five DNA polymerases exist in E. coli, called Pol I through Pol V. Pol
III is the principal enzyme responsible for synthesis of new DNA, and it is a
multiple-subunit enzyme.
DNA polymerases I and II are involved in proofreading and repair
processes.
All DNA synthesis requires an RNA primer.
Related Topics