Chapter: Biochemistry: Biosynthesis of Nucleic Acids: Replication
Eukaryotic DNA Replication
Eukaryotic DNA Replication
Our
understanding of replication in eukaryotes is not as extensive as that in
prokaryotes, owing to the higher level of complexity in eukaryotes and the
consequent difficulty in studying the processes. Even though many of the
principles are the same, eukaryotic replication is more complicated in three
basic ways: there are multiple origins of replication, the timing must be
controlled to that of cell divisions, and more proteins and enzymes are
involved.
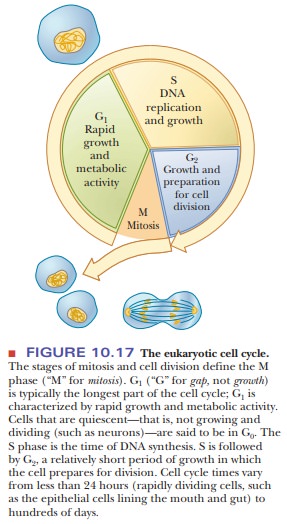
In a
human cell, a few billion base pairs of DNA must be replicated once, and only
once, per cell cycle. Cell growth and division are divided into phases-M, G1, S, and
G2 (Figure 10.17). DNA replication takes place during a few hours in
the S phase, and pathways exist to make sure that the DNA is replicated only
once per cycle. Eukaryotic chromosomes accomplish this DNA synthesis by having
replication begin at multiple origins of replication, also called replicators. These are specific DNA
sequences that are usually between gene sequences. An average human chromosome
may have several hundred replicators. The zones where replication is proceeding
are called replicons, and the size
of these varies with the species. In higher mammals, replicons may span 500 to
50,000 base pairs.
How is replication tied to cell division?
The
best-understood model for control of eukaryotic replication is from yeast cells
(Figure 10.18). Only chromosomes from cells that have reached the G1 phase
are competent to initiate DNA replication. Many proteins are involved in the
control of replication and its link to the cell cycle. As usual, these proteins
are usually given an abbreviation that makes them easier to say, but more
difficult for the uninitiated to comprehend at first glance. The first proteins
involved are seen during a window of opportunity that occurs between the early
and late G1 phase (see Figure 10.18, top).
Replication is initiated by a multisubunit protein called theorigin recognitioncomplex (ORC), which
binds to the origin of replication. This protein complexappears to be bound to
the DNA throughout the cell cycle, but it serves as an attachment site for
several proteins that help control replication. The next protein to bind is an
activation factor called the replication
activator protein (RAP). After the activator protein is bound, replication licensing factors (RLFs)
can bind. Yeast contains at least six different RLFs. They get their name from
the fact that replication cannot proceed until they are bound. One of the keys
to linking replication to cell division is that some of the RLF proteins have
been found to be cytosolic. Thus, they have access to the chromosome only when
the nuclear membrane dissolves during mitosis. Until they are bound, replication
cannot occur. After RLFs bind, the DNA is then competent for replication. The
combination of the DNA, ORC, RAP, and RLFs constitutes what researchers call
the pre-replication complex (pre-RC).
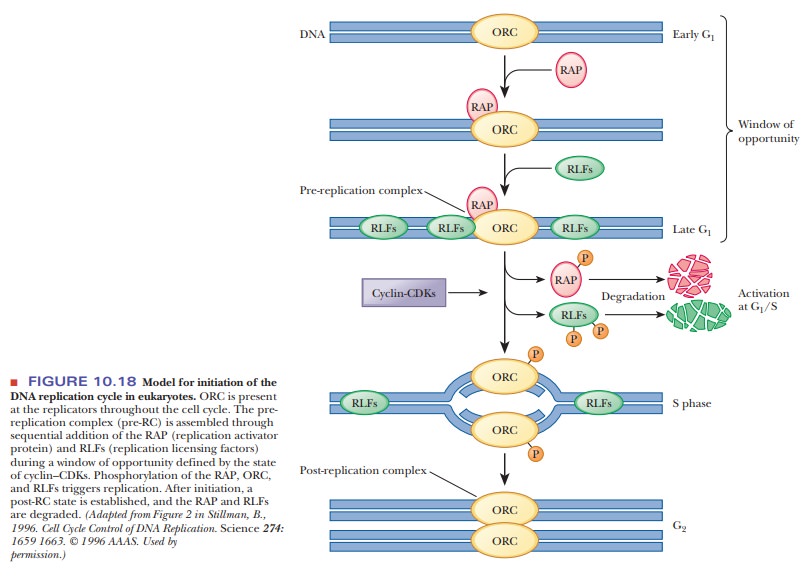
The next step involves other proteins and protein kinases. We learned that many processes are controlled by kinases phosphorylating target proteins.
One of the great discoveries in this field was the existence of cyclins, which are proteins that are
produced in one part of a cell cycle and degraded in another. Cyclins are able
to combine with specific protein kinases, called cyclin-dependent protein kinases (CDKs). When these cyclins combine
with CDKs, theycan activate DNA replication and also block reassembly of a
pre-RC after initia-tion. The state of activity of the CDKs and the cyclins
determines the window of opportunity for DNA synthesis. Cyclin–CDK complexes
phosphorylate sites on the RAP, the RLFs, and the ORC itself. Once
phosphorylated, the RAP dissoci-ates from the pre-RC, as do the RLFs. Once
phosphorylated and released, the RAP and the RLFs are degraded (Figure 10.18, middle). Thus, the activation of
cyclin–CDKs serves both to initiate DNA replication and to prevent formation of
another pre-RC. In the G2 phase, the DNA has been
replicated. During mitosis, the DNA is separated into the daughter cells. At
the same time, the dissolved nuclear membrane allows entrance of the licensing
factors that are produced in the cytosol so that each daughter cell can initiate
a new round of replication.
Eukaryotic DNA Polymerases
At least
15 different polymerases are present in eukaryotes, of which 5 have been
studied more extensively (Table 10.4). The use of animals rather than plants
for study avoids the complication of any DNA synthesis in chloroplasts. The
five best-studied polymerases are called α, β, γ, δ, and ε. The α, β, δ, and ε enzymes are found in the nucleus, and the γ form occurs in mitochondria.
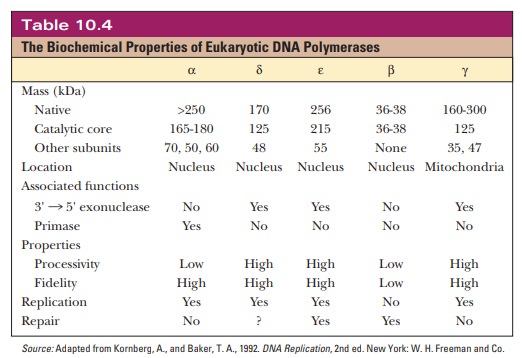
How are eukaryotic polymerases related to prokaryotic ones?
Polymerase
α was the
first discovered, and it has the most subunits. It also has the ability to make
primers, but it lacks a 3' - > 5' proofreading activity and has low
processivity. After making the RNA primer, Pol α adds about 20 nucleotides and is then replaced
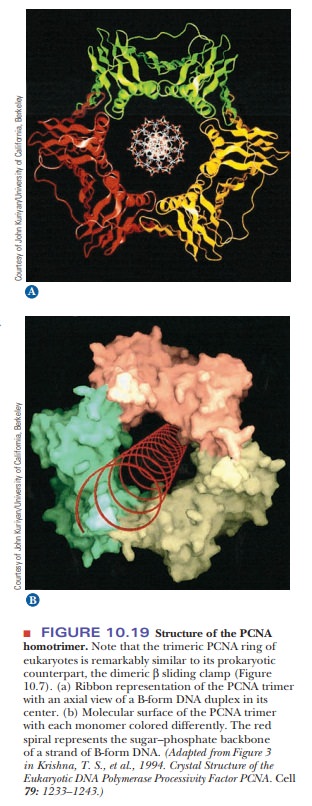
by Pol δ and ε. Polymerase δ is the principal DNA
polymerase in eukaryotes. It interacts with a special protein called PCNA (for proliferating cell nuclearantigen). PCNA is the eukaryotic
equivalent of the part of Pol III that functionsas a sliding clamp (β). It is a trimer of three
identical proteins that surround the DNA (Figure 10.19). The role of DNA
polymerase ε is less
clear. It may replace polymerase δ in lagging strand synthesis. DNA polymerase β appears to be a repair
enzyme. DNA polymerase γ carries out DNA replication in mitochondria.
Several of the
DNA polymerases isolated from animals lack exonuclease activity (the α and β enzymes). In this regard, the animal enzymes
differ from prokaryotic DNA polymerases. Separate exonucleolytic enzymes exist
in animal cells.
The Eukaryotic Replication Fork
The general features of DNA replication in eukaryotes are similar to those in prokaryotes. Table 10.5 summarizes the differences. As with prokaryotes, DNA replication in eukaryotes is semiconservative. There is a leading strand with continuous synthesis in the 5' - > 3' direction and a lagging strand with discontinuous synthesis in the 5' - > 3' direction. An RNA primer is formed by a specific enzyme in eukaryotic DNA replication, as is the case with prokaryotes, but in this case the primase activity is associated with Pol α.
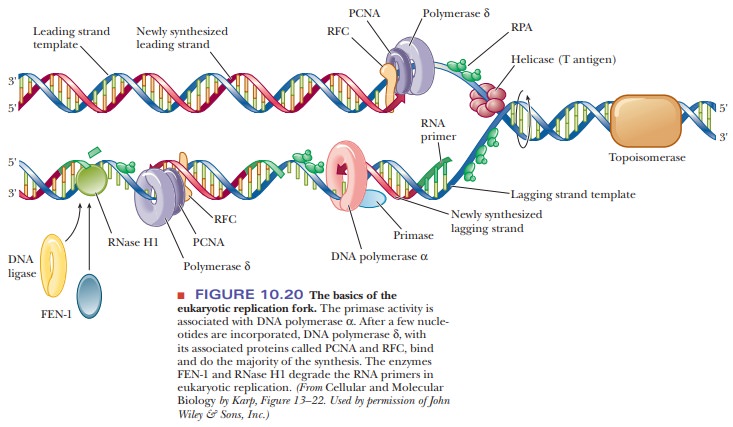
The structures involved
at the eukaryotic replication fork are shown in Figure 10.20. The formation of
Okazaki fragments (typically 150 to 200 nucleotides long in eukaryotes) is
initiated by Pol α. After
the RNA primer is made and a few nucleotides are added by Pol α, the polymerase dissociates
and is replaced by Pol δ and its attached PCNA protein. Another
protein, called RFC (replication
factor C), is involved in attaching PCNA to Pol δ. The RNA primer is eventually degraded, but,
in the case of eukaryotes, the polymerases do not have the 5'3 3' exonuclease
activity to do it. Instead, separate enzymes, FEN-1 and RNase H1, degrade the
RNA. Continued movement of Pol δ fills in the gaps made by primer removal. As
with prokaryotic replication, topoisomerases relieve the torsional strain from
unwinding the helix, and a single-stranded binding protein, called RPA,
protects the DNA from degradation. Finally, DNA ligase seals the nicks that
separate the fragments.
Another important difference between DNA replication in prokaryotes and in eukaryotes is that prokaryotic DNA is not complexed to histones, as is eukaryotic DNA. Histone biosynthesis occurs at the same time and at the same rate as DNA biosynthesis. In eukaryotic replication, histones are associated with DNA as it is formed. An important aspect of DNA replication in eukaryotes, specifically affecting humans.
Summary
Replication in eukaryotes
follows the same general outline as replication in prokaryotes, with the most
important difference being the presence of histone proteins complexed to
eukaryotic DNA.
Different proteins are used,
and the system is more complex than it is in prokaryotes. Replication is
controlled so that it occurs only once during a cell-division cycle, during the
S phase.
Five
different DNA polymerases are present in eukaryotes: α, β, γ, δ, and ε. Polymerase δ is the principal synthesizer of DNA and is the
equivalentof Pol III in prokaryotes.
Related Topics