Chapter: Biochemistry: Biosynthesis of Nucleic Acids: Replication
Proteins Required for DNA Replication
Proteins Required for DNA
Replication
Two
questions arise in separating the two strands of the original DNA so that it
can be replicated. The first is how to achieve continuous unwinding of the
double helix. This question is complicated by the fact that prokaryotic DNA
exists in a supercoiled, closed-circular form (see “Tertiary Structure of DNA:
Supercoiling”). The second related question is how to protect single-stranded
stretches of DNA that are exposed to intracellular nucleases as a result of the
unwinding.
Supercoiling and Replication
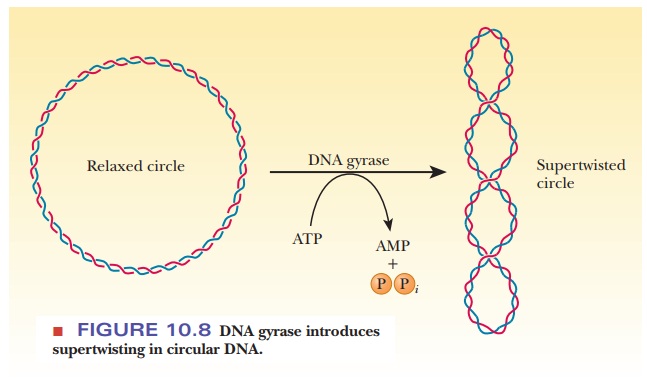
An
enzyme called DNA gyrase (class II
topoisomerase) catalyzes the conversion of relaxed, circular DNA with a nick in
one strand to the supercoiled form with the nick sealed that is found in normal
prokaryotic DNA (Figure 10.8). A slight unwinding of the helix before the nick
is sealed introduces the supercoiling. The energy required for the process is
supplied by the hydrolysis of ATP. Some evidence exists that DNA gyrase causes
a double-strand break in DNA in the process of converting the relaxed, circular
form to the supercoiled form.
How does replication work with supercoiled DNA?
In
replication, the role of the gyrase is somewhat different. The prokaryotic DNA
is negatively supercoiled in its natural state; however, opening the helix
during replication would introduce positive supercoils ahead of the replication
fork. To see this phenomenon for yourself, look for an old phone with a coiled
cord and try straightening out a section of the cord. You will be able to see
the result in the coils ahead. If the replication fork continued to move, the
torsional strain of the positive supercoils would eventually make further
replication impossible. DNA gyrase fights these positive supercoils by putting
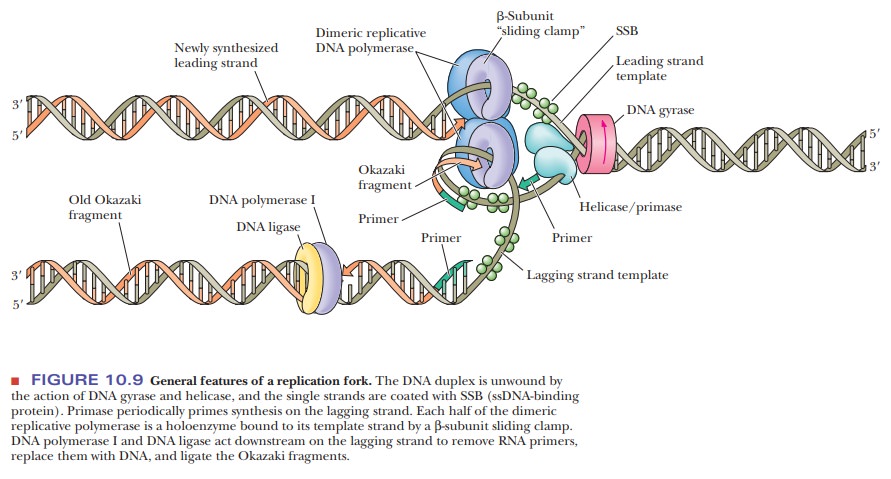
negative supercoils ahead of the replication fork (Figure 10.9). A
helix-destabilizing protein, called a helicase,
promotes unwinding by binding at the replication fork. A number ofhelicases
are known, including the DnaB protein
and the rep protein.
How is single-stranded DNA protected long enough for replication?
Single-stranded regions of
DNA are very susceptible to degradation by nucleases.
If left unchecked, this would make it very difficult to complete replication before DNA damage would occur.
Another protein, called the single-strand bindingprotein (SSB), stabilizes the single-stranded
regions by binding tightly to theseportions of the molecule. The presence of
this DNA-binding protein protects the single-stranded regions from hydrolysis
by the nucleases.
The Primase Reaction
One of
the great surprises in studies of DNA replication was the discovery that RNA serves as a primer in DNA replication. In
retrospect, it is not surprising atall, because RNA can be formed de novo
without a primer, even though DNA synthesis requires a primer. This finding
lends support to theories of the origin of life in which RNA, rather than DNA,
was the original genetic material. The fact that RNA has been shown to have
catalytic ability in several cases has added support to that theory. A primer in
DNA replication must have a free 3'-hydroxyl to which the growing chain can
attach, and both RNA and DNA can provide this group. The primer activity of RNA
was first observed in vivo. In some of the original in vitro experiments, DNA
was used as a primer because a primer consisting of DNA was expected. Living
organisms are, of course, far more complex than isolated molecular systems and,
as a result, can be full of surprises for researchers.
Where does the primer come from?
It has
subsequently been found that a separate enzyme, called primase, is responsible for copying a short stretch of the DNA
template strand to produce the RNA primer sequence. The first primase was
discovered in E. coli. The enzyme
consists of a single polypeptide chain, with a molecular weight of about
60,000. There are 50 to 100 molecules of primase in a typical E. coli cell. The primer and the protein
molecules at the replication fork constitute the primosome. The general features of DNA replication, including the
use of anRNA primer, appear to be common to all prokaryotes (Figure 10.9).
Synthesis and Linking of New DNA Strands
The
synthesis of two new strands of DNA is begun by DNA polymerase III. The newly
formed DNA is linked to the 3'-hydroxyl of the RNA primer, and synthesis proceeds
from the 5' end to the 3' end on both the leading and the lagging strands. Two
molecules of Pol III, one for the leading strand and one for the lagging
strand, are physically linked to the primosome.
The resulting multiprotein complex is called the replisome. As the replication fork moves, the RNA primer is removed
by polymerase I, using its exonuclease activity. The primer is replaced by
deoxynucleotides, also by DNA polymerase I, using its polymerase activity. (The
removal of the RNA primer and its replacement with the missing portions of the
newly formed DNA strand by polymerase I are the repair function we mentioned
earlier.) None of the DNA polymerases can seal the nicks that remain; DNA
ligase is the enzyme responsible for the final linking of the new strand. Table
10.3 summarizes the main points of DNA replication in prokaryotes.
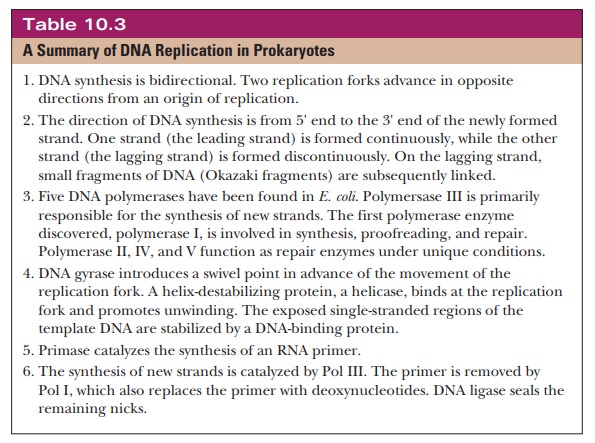
Summary
Besides the DNA polymerases
themselves, many other proteins are involved in replication. DNA gyrase induces
negative supercoils in the DNA to compensate for the positive supercoils that
would form because of strand separation, and helicase induces strand
separation. Single-stranded binding proteins protect the single-stranded
regions from nucleases.
Primase primes the synthesis of the lagging
strand by the formation of primers, and DNA ligase links pieces of newly formed
DNA together.
The primer and the proteins at the replication
fork are called the primosome.
The
entire complex, including the DNA polymerases, is called the replisome.
Related Topics