Chapter: Biochemistry: Biosynthesis of Nucleic Acids: Replication
Proofreading and Repair
DNA replication takes place only once each
generation in each cell, unlike other processes, such as RNA and protein
synthesis, which occur many times. It is essential that the fidelity of the
replication process be as high as possible to prevent mutations, which are errors in replication. Mutations are
frequently harmful, even lethal, to organisms. Nature has devised several ways
to ensure that the base sequence of DNA is copied faithfully.
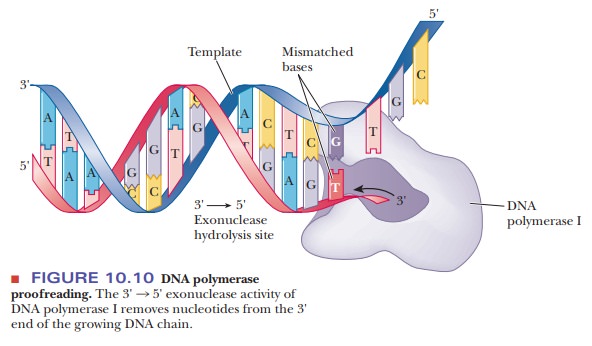
Errors in replication occur spontaneously only
once in every 109 to 1010 base pairs. Proofreading refers to the removal of
incorrect nucleotides immediately after they are added to the growing DNA
during the replication process. DNA polymerase I has three active sites, as
demonstrated by Hans Klenow. Pol I can be cleaved into two major fragments. One
of them (the Klenow fragment) contains the polymerase activity and the
proofreading activity. The other con-tains the 5' - > 3' repair activity.
Figure 10.10 shows the proofreading activity of Pol I. Errors in hydrogen
bonding lead to the incorporation of an incorrect nucleotide into a growing DNA
chain once in every 104 to 105 base pairs. DNA polymerase
I uses its 3' exonuclease activity to remove the incorrect nucleotide.
Replication resumes when the correct nucleotide is added, also by DNA
poly-merase I. Although the specificity of hydrogen-bonded base pairing
accounts for one error in every 104 to 105 base pairs,
the proofreading function of DNA polymerase improves the fidelity of
replication to one error in every 109 to 1010 base pairs.
How does proofreading improve replication fidelity?
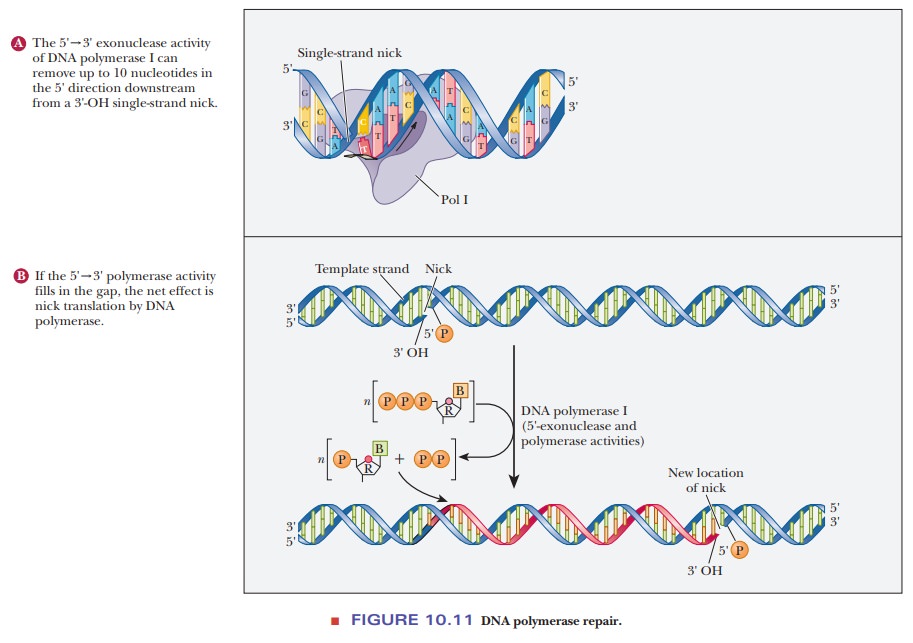
During
replication, a cut-and-patch process
catalyzed by polymerase I takes place. The cutting is the removal of the RNA
primer by the 5' exonuclease function of the polymerase, and the patching is
the incorporation of the required deoxynucleotides by the polymerase function
of the same enzyme. Note that this part of the process takes place after
polymerase III has produced the new polynucleotide chain. Existing DNA can also
be repaired by polymerase I, using the cut-and-patch method, if one or more
bases have been damaged by an external agent, or if a mismatch was missed by
the proofreading activity. DNA polymerase I is able to use its 5' - > 3'
exonuclease activity to remove RNA primers or DNA mistakes as it moves along
the DNA. It then fills in behind it with its polymerase activity. This process
is called nick translation (Figure
10.11). In addition to experiencing those spontaneous mutations caused by
misreading the genetic code, organisms are frequently exposed to mutagens, agents that produce
mutations.
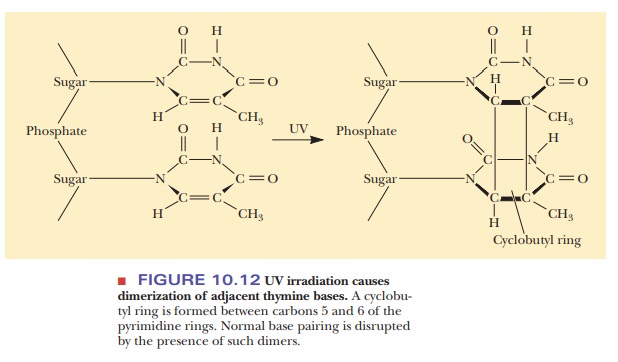
Common mutagens include ultraviolet light, ionizing
radiation (radioactivity), and various chemical agents, all of which lead to
changes in DNA over and above those produced by spontaneous mutation. The most
common effect of ultraviolet light is the creation of pyrimidine dimers (Figure
10.12). The π
electrons from two carbons on each of two pyrimidines form a cyclobutyl ring,
which distorts the normal shape of the DNA and interferes with replication and
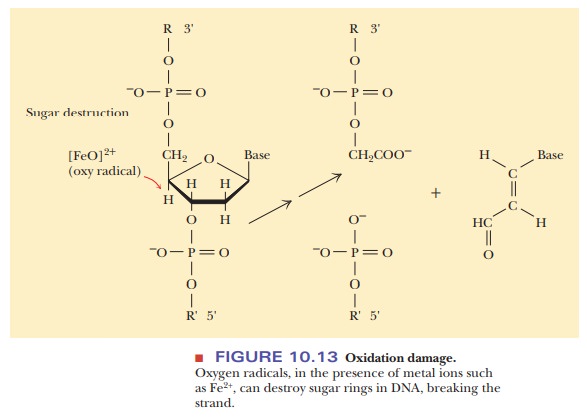
transcription. Chemical damage, which is often caused by free radicals (Figure
10.13), can lead to a break in the phosphodiester backbone of the DNA strand.
This is one of the primary reasons that antioxidants are so popular as dietary
supplements these days.
When
damage has managed to escape the normal exonuclease activities of DNA
polymerases I and III, prokaryotes have a variety of other repair mecha-nisms
at their disposal. In mismatch repair,
enzymes recognize that two bases are incorrectly paired. The area with the
mismatch is removed, and DNA polymerases replicate the area again. If there is
a mismatch, the challenge for the repair system is to know which of the two
strands is the correct one. This is possible only because prokaryotes alter
their DNA at certain locations by modifying bases with added methyl groups.
This methylation occurs shortly
after replication. Thus, immediately after replication, there is a window of
opportunity for the mismatch-repair system. Figure 10.14 shows
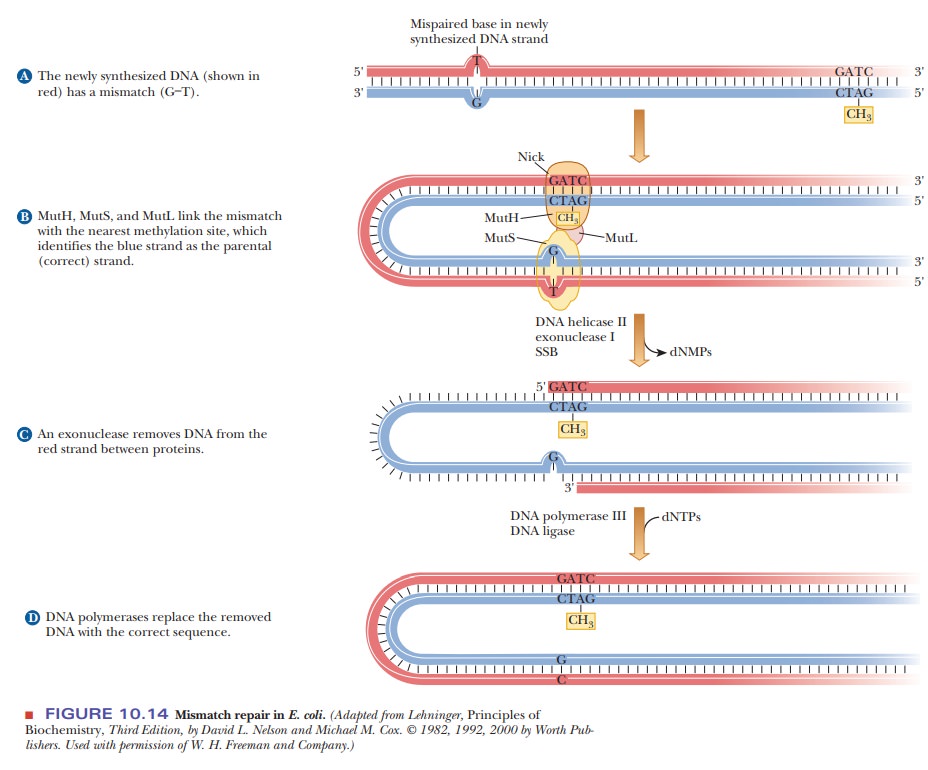
Assume that a bacterial species methylates adenines that are part of a
unique sequence. Originally, both parental strands are methylated. When the DNA
is replicated, a mistake is made, and a T is placed opposite a G (Figure
10.14a). Because the parental strand contained methylated adenines, the enzymes
can distinguish the parental strand from the newly synthesized daughter strand
without the modified bases. Thus, the T is the mistake and not the G. Several
proteins and enzymes are then involved in the repair process. MutH, MutS, and MutL form a loop between the mistake and a methylation site.DNA
helicase II helps unwind the DNA. Exonuclease
I removes the section of DNA containing the mistake (Figure 10.14b).
Single-stranded binding proteins protect the template (blue) strand from
degradation. DNA polymerase III then fills in the missing piece (Figure
10.14c).
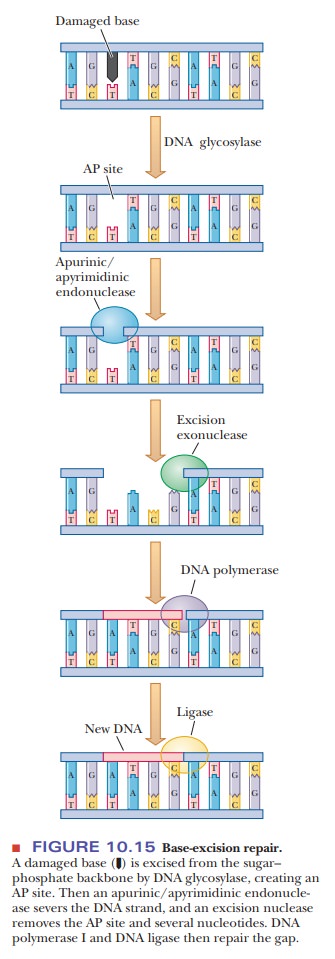
Another
repair system is called base-excision
repair (Figure 10.15). A base that has been damaged by oxidation or
chemical modification is removed by DNA
glycosylase, leaving an AP site,
so called because it is apurinic or apy-rimidinic (without purine or
pyrimidine). An AP endonuclease then
removes the sugar and phosphate from the nucleotide. An excision exonuclease then removes several more bases. Finally, DNA
polymerase I fills in the gap, and DNA ligase seals the phosphodiester
backbone.
Nucleotide-excision repair is
common for DNA lesions caused by ultravioletor chemical means, which often lead
to deformed DNA structures. Figure 10.16 demonstrates how a large section of
DNA containing the lesion is removed by ABC
excinuclease. DNA polymerase I and DNA ligase then work to fill in the gap.This
type of repair is also the most common repair for ultraviolet damage in
mammals. Defects in DNA repair mechanisms can have drastic consequences. One of
the most remarkable examples is the disease xeroderma
pigmentosum. Affected individuals develop numerous skin cancers at an early
age because

The
endonuclease that nicks the damaged portion of the DNA is prob-ably the missing
enzyme. The repair enzyme that recognizes the lesion has been named XPA protein
after the disease. The cancerous lesions eventually spread throughout the body,
causing death.
Summary
Bases would be paired incorrectly during DNA synthesis about once
for every 104 to 105 base pairs unless there were a
mechanism to increase fidelity.
Because
of proofreading and repair, the number of incorrect bases is reduced to one in
109 to 1010.
Proofreading is the process by which DNA
polymerase I removes incor-rectly paired bases immediately after they are added
to the growing chain.
Related Topics