Chapter: Biotechnology: Protein Structure And Engineering
Protein Finger printing- Peptide Mapping
Protein Finger printing- Peptide Mapping
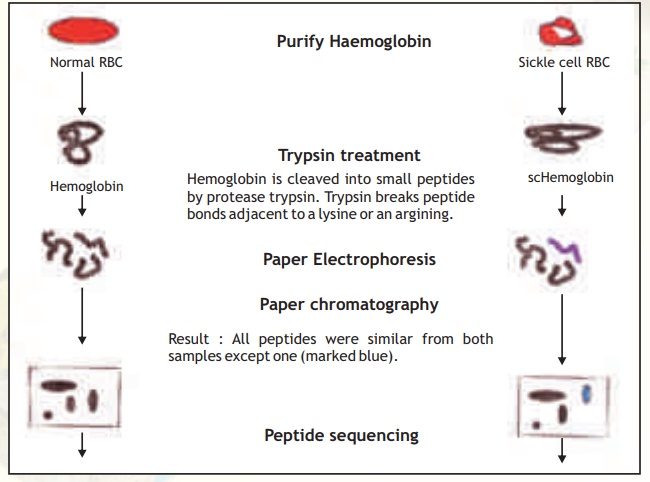
This technique involves the generation and 2-D analysis of peptides from a protein. Each protein has a unique peptide map (2-D analysis) and hence serves as a fingerprint for the protein. The steps involved in generating a peptide map/fingerprint are as follows(Fig. 6).
1. Pure Hb and scHb are taken separately into test tubes.
2. The Hb and scHb are digested with the proteolytic enzyme trypsin which cleaves the protein after basic amino acid residues Arg and Lys.
3. Two separate strips of Whatman filter paper are spotted with Hb and scHb tryptic peptides and the peptides allowed to separate using the technique of paper electrophoresis at pH 2.0. Highly charged peptides will migrate more towards the anode/cathode.
4. The paper strips are dried, attached to larger squares of Whatman paper and chromatographed at right angles to the electrophoretic direction using a solvent system Butanol: Water:Acetic acid. In such a system peptides will separate based on
their partition coefficient between the solvent and paper which is dependant on the relative hydrophobicity of the peptides. More hydrophobic peptides will move with the solvent to longer distances.
5. The chromatograms are dried and stained with a suitable visualisation reagent like Ninhydrin wherein peptide containing regions appear as orange yellow spots.
6. The peptide map for Hb and scHb are compared and it was found that one peptide was differently placed in the scHb map.
7. On examining this peptide and determining its amino acid sequence, Ingram found that it had a valine substitution for glutamic acid in the peptide.
The single substitution of valine for glutamic acid (val/glu are at the 6th position of the haemoglobin beta chain) dramatically changes the structure of scHb making it form fibres within the RBC resulting in the deformation of the cell (sickling). Since the disease was due to a molecular alteration the term molecular disease was applied.
Peptide mapping became a useful technique to compare similar proteins from different sources. Slowly the information became too vast and computers were used to store this data into databases so that homology searches could be made. The protein fingerprinting data has been further augmented with new databases containing 2-D electrophoresis patterns of entire proteins from a given cell type, a technique developed by O'Farrel.
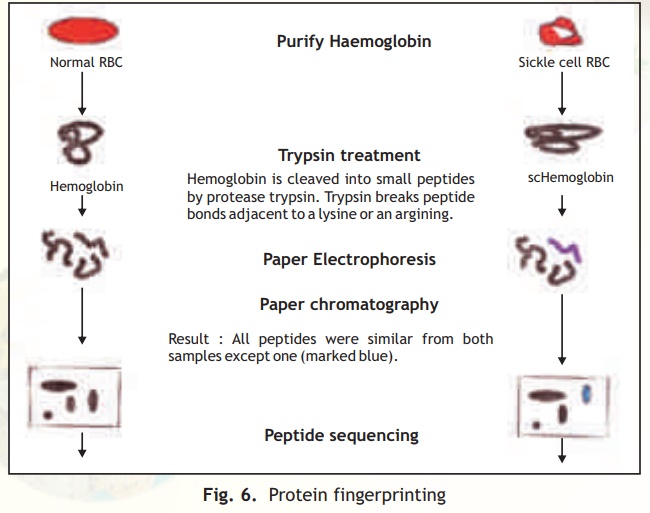
Fig. 6. Protein fingerprinting
Related Topics