Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Viral Multiplication
Problems of Producing mRNA
THE PROBLEMS OF PRODUCING mRNA
From Genome to mRNA
An essential step in every virus infection is the production of virus-specific mRNAs that program the cellular ribosomes to synthesize viral proteins. Besides the structural pro-teins of the virion, viruses must direct the synthesis of enzymes and other specialized pro-teins required for genome replication, gene expression, and virus assembly and release. The production of the first viral mRNAs at the beginning of the infection is a crucial step in the takeover of the cell by the virus.
For some viruses, the presentation of mRNA to the cellular ribosomes poses no prob-lems. Thus, the genomes of most DNA viruses are transcribed by the host DNA-dependent RNA polymerase to yield the viral mRNAs. The ( )-strand RNA viruses, such as the picornaviruses, the togaviruses, and the coronaviruses, possess genomes that can be used directly as mRNAs and are translated (at least partially, as discussed later) immediately on entry into the cytoplasm of the cell.
However, for many viruses, the production of mRNA starting from the genome is not so straightforward. The fact that poxviruses replicate in the cytoplasm means that the cel-lular RNA polymerase is not available to transcribe the DNA genome. Moreover, no cellular machinery exists that can use either single-stranded or double-stranded RNA as a template to synthesize mRNA. Therefore, the poxviruses and those viruses that utilize an RNA template to make mRNAs must provide their own transcription machinery to pro-duce the viral mRNAs at the beginning of the infection process. This feat is accomplished by synthesizing the transcriptases in the later stages of viral development in the previous host cell and packaging the enzymes into the virions, where they remain associated with the genome as the virus enters the new cell and uncoats. In general, the presence of a transcriptase in virions is indicative that the host cell is unable to use the viral genome as mRNA or as a template to synthesize mRNA. At later times in the infection, any special enzymatic machinery required by the virus and not initially present in the cell, can be supplied among the proteins translated from the first mRNA molecules.
The pathways for the synthesis of mRNA by the major virus groups are summarized in Figure 6 – 5 and related to the structure of viral genomes. The polarity of mRNA is des-ignated as (+) and the polarity of polynucleotide chains complementary to mRNA as (-). The black arrows denote synthetic steps for which host cells provide the required enzymes, whereas the colored arrows indicate synthetic steps that must be carried out by virus-encoded enzymes. Several additional points should be emphasized. The par-voviruses and some phages have single-stranded DNA genomes. Although the RNA po-lymerase of the cell requires double-stranded DNA as a template, these viruses need not
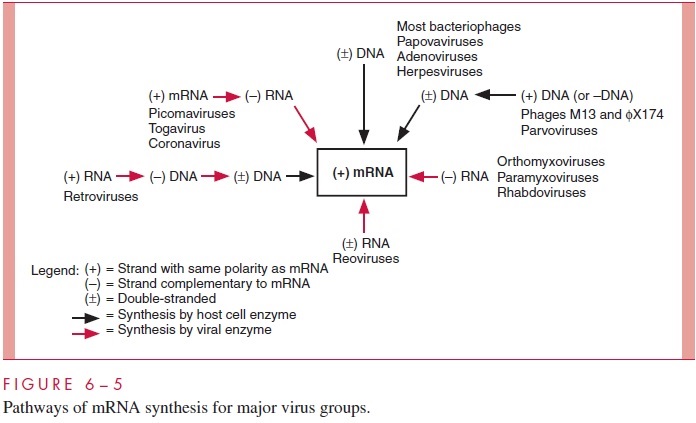
carry special enzymes in their virions because host cell DNA polymerases can convert the genomes into double-stranded DNA. Note that the production of more mRNA by the pi-cornaviruses and similar (+)-strand RNA viruses requires the synthesis of an intermedi-ate (-)-strand RNA template. The enzyme required for this process is produced by translation of the genome RNA early in infection.
The retroviruses are a special class of (+)-strand RNA viruses. Although their genomes are the same polarity as mRNA and could in principle serve as mRNAs early af-ter infection, their replication scheme apparently precludes this. Instead, the RNA genomes of these viruses are copied into (-)DNA strands by an enzyme carried within the virion called reverse transcriptase. The (-)DNA strands are subsequently converted by the same enzyme to double-stranded DNA in a reaction that requires the degradation of the original genomic RNA by the RNase H activity of the reverse transcriptase. The DNA product of reverse transcription is integrated into the host cell DNA and ultimately transcribed by the host RNA polymerase to complete the replication cycle as well as pro-duce viral mRNA. For example, the replication of the hepatitis B DNA genome is mecha-nistically similar to that of a retrovirus. Thus, the viral DNA is transcribed to produce a single-stranded RNA, which in turn is reverse transcribed to produce the progeny viral DNA that is encapsidated into virions.
The Monocistronic mRNA Rule in Animal Cells
The ribosome requires input of information in the form of mRNA. For a viral mRNA to be recognized by the ribosome, its production must conform to the rules of structure that govern the synthesis of the cellular mRNAs. Prokaryotic mRNA is relatively simple and can be polycistronic, which means it can contain the information for several proteins. Each cistron or coding region is translated independently beginning from its own ribo-some binding site.
Eukaryotic mRNAs are structurally more complex, containing special 5’ -cap and 3’ -poly(A) attachments. In addition, their synthesis often involves removal of internal se-quences by a process called splicing. Most importantly, virtually all eukaryotic mRNAs are monocistronic. Accordingly, eukaryotic translation is initiated by the binding of a ri-bosome to the 5 -cap, followed by movement of the ribosome along the DNA until the first AUG initiation codon is encountered. The corollary to this first AUG rule is that eu-karyotic ribosomes, unlike prokaryotic ribosomes, generally cannot initiate translation at internal sites on a mRNA. To conform to the monocistronic mRNA, most animal viruses produce mRNAs that are translated to yield only a single polypeptide chain following ini-tiation near the 5’ end of the mRNA.
Because most DNA animal viruses replicate in the nucleus, they adhere to the mono-cistronic mRNA rule either by having a promoter precede each gene or by programming the transcription of precursor RNAs that are processed by nuclear splicing enzymes into monocistronic mRNAs. The virion transcriptase of the cytoplasmic poxviruses apparently must synthesize monocistronic mRNAs by initiation of transcription in front of each gene.
RNA-containing animal viruses have evolved three different strategies to circumvent or conform to the monocistronic mRNA rule. The simplest strategy involves having a segmented genome. For the most part, each genome segment of the orthomyxoviruses and the reoviruses corresponds to a single gene; therefore, the mRNA transcribed from a given segment constitutes a monocistronic mRNA. Unlike most RNA viruses, the or-thomyxovirus virus influenza A replicates in the nucleus, and some of its monocistronic mRNAs are produced by splicing of precursor RNAs by host cell enzymes. Moreover, orthomyxoviruses use small 5’ RNA fragments derived from host cell pre-mRNAs found in the nucleus to prime the synthesis of their own mRNAs.
A second solution to the monocistronic mRNA rule is very similar to the strategy em-ployed by cells and the DNA viruses. The paramyxoviruses, togaviruses, rhabdoviruses, filoviruses, bunyaviruses, arenaviruses, and coronaviruses synthesize monocistronic mRNAs by initiating the synthesis of each mRNA at the beginning of a gene. In most cases, the tran-scriptase terminates mRNA synthesis at the end of the gene so that each message corre-sponds to a single gene. For coronaviruses, RNA synthesis initiates at the beginning of each gene and continues to the end of the genome so that a nested set of mRNAs is produced. However, each mRNA is functionally monocistronic and is translated to produce only the protein encoded near its 5’ end.
The picornaviruses have evolved yet a third strategy to deal with the monocistronic mRNA requirement (Fig 6–6). The (+)-strand genome contains just a single ribosome binding site near the 5’ end. It is translated into one long polypeptide chain called a polyprotein, which is subsequently broken into the final set of protein products by a series of proteolytic cleavages. Most of the required protease activities reside within the polyprotein itself.
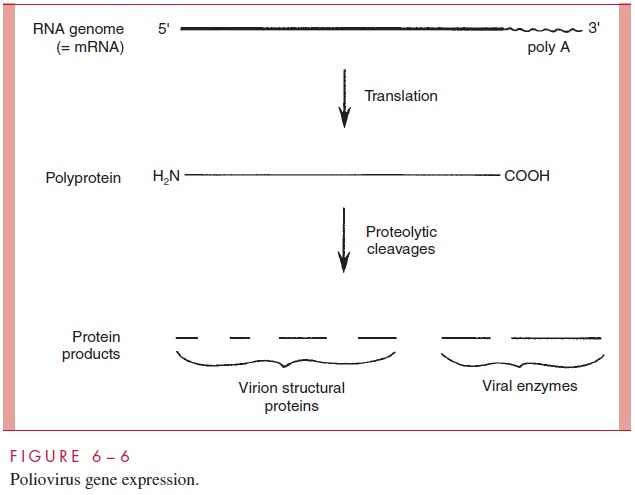
Several viruses use more than one of these strategies to conform to the monocistronic mRNA rule. For example, retroviruses, togaviruses, arenaviruses, and bunyaviruses syn-thesize multiple mRNAs, each one coding for a polyprotein that is subsequently cleaved into the individual protein molecules.
Related Topics