Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Viral Multiplication
Assembly of Naked Capsid Viruses and Nucleocapsids
ASSEMBLY OF NAKED CAPSID VIRUSES AND NUCLEOCAPSIDS
The process of enclosing the viral genome in a protein capsid is called assembly or encapsidation. Four general principles govern the construction of capsids and nucleocapsids.First, the process generally involves self-assembly of the component parts. Second, assem-bly is stepwise and ordered. Third, individual protein structural subunits or protomers are usually preformed into capsomeres in preparation for the final assembly process. Fourth, assembly often initiates at a particular locus on the genome called a packaging site.
Viruses With Helical Symmetry
The assembly of the cylindrically shaped tobacco mosaic virus (TMV) has been extensively studied and provides a model for the construction of helical capsids and nucleocapsids. For TMV, doughnut-shaped disks containing a number of individual structural subunits are pre-formed and added stepwise to the growing structure. Elongation occurs in both directions from a specific packaging site on the single-stranded viral RNA (Fig 6 – 9). The addition of

each disk involves an interaction between the protein subunits of the disk and the genome RNA. The nature of this interaction is such that the assembly process ceases when the ends of the RNA are reached. The structural subunits as well as the RNA trace out a helical path in the final virus particle.
The basic design features worked out for TMV probably apply in general to the as-sembly of the nucleocapsids of enveloped viruses. Thus, it is likely that the individual protein subunits are intimately associated with the RNA and that the nucleoprotein com-plexes are assembled by the stepwise addition of protein subunits or complexes of sub-units. For influenza and the other helical viruses with segmented genomes, the various genome segments are assembled into nucleocapsids independently and then brought to-gether during virion assembly by a mechanism that is as yet poorly understood. It is no-table that virtually all of the animal RNA viruses with helical symmetry are enveloped.
Viruses with Cubic Symmetry
For both phage and animal viruses, icosahedral capsids are generally preassembled and the nucleic acid genomes, usually complexed with condensing proteins, are threaded into the empty structures. Construction of the hollow capsids appears to occur by a self-assembly process, sometimes aided by other proteins. The stepwise assembly of compo-nents involves the initial aggregation of structural subunits into pentamers and hexamers, followed by the condensation of these capsomeres to form the empty capsid. In some cases, it appears that a small complex of capsid proteins associates specifically with the viral genome and nucleates the assembly of the complete capsid around the genome.
The morphogenesis of a complex bacteriophage such as T4 involves the prefabrica-tion of each of the major substructures by a separate pathway, followed by the ordered and sequential construction of the final particle from its component parts (Fig 6 – 10). An intermediate in the assembly of a bacteriophage head is an empty structure containing an internal protein network that is removed prior to insertion of the nucleic acid. The con-stituents of this network are often appropriately referred to as scaffolding proteins, which apparently provide the lattice necessary to hold the capsomeres in position during the early stages of head assembly.
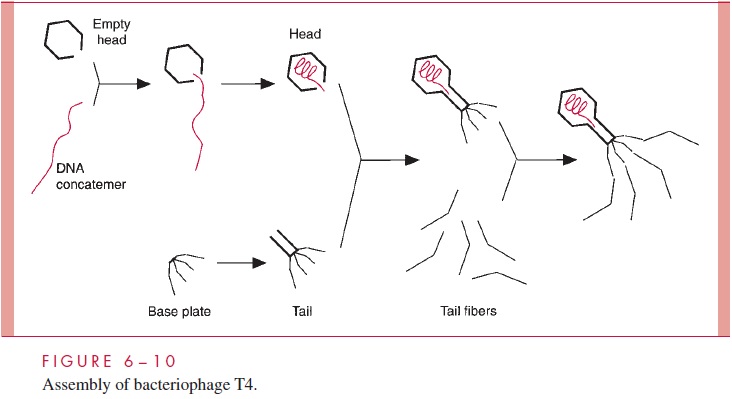
For many DNA bacteriophages and the herpesviruses, the products of replication are long linear DNA molecules called concatemers, which are made up of tandem head-to-tail repeats of genome-size units. During the threading of the DNA into the preformed capsids, these concatemers are cleaved by virus-encoded nucleases to generate genome-size pieces.
There are two mechanisms for determining the correct sites for nuclease cleavage during packaging of a concatemer. Bacteriophage l and the herpesviruses typify one type of mechanism in which the enzyme that makes the cuts is a sequence-specific nu-clease. The enzyme sits poised at the orifice of the capsid as the DNA is being threaded into the capsid, and just before the specific cut site enters, the DNA is cleaved. For bacteriophage l, the breaks are made in opposite strands, 12 bp apart, to generate the cohe-sive ends. Bacteriophages T4 and P1 are examples of bacterial viruses that illustrate the second mechanism. For these phages, the nuclease does not recognize a particular DNA sequence, but instead cuts the concatemer when the capsid is full. Because the head of the bacteriophage can accommodate slightly more than one genome equivalent of DNA and packaging can begin anywhere on the DNA, the “headful” mechanism produces genomes that are terminally redundant (the same sequence is found at both ends) and circularly permuted. The nonspecific packaging with respect to DNA sequence explains why bacteriophage P1 is capable of incorporating host DNA into phage particles, thereby promoting generalized transduction . Bacteriophage T4 does not carry out generalized transduction, because the bacterial DNA is completely degraded to nucleotides early in infection.
Related Topics