Chapter: Genetics and Molecular Biology: Advanced Genetic Engineering
Polymerase Chain Reaction
Polymerase Chain Reaction
A method has been devised with such remarkable
sensitivity that it can detect a single molecule of a specific sequence.
Furthermore, the single molecule can be detected in the presence of a 106 or
greater excess of other sequences. This method is called the polymerase chain
reaction or PCR. The polymerase chain reaction is also useful for studying
specific genes or sequences. For example, it permits the sequencing in a day or
two of a stretch of several hundred nucleotides with the starting point being a
small sample of blood. No cloning is required for the sequencing. Such
sensitivity permits the rapid characterization of the basis of mutations or of
genetic defects. This extraordinary sensitivity also provides a sensitive test
for the presence of a virus like HIV. Again a sample of blood can be taken and
the assay can detect the presence of one copy of the single virus sequence in
100,000 cells. The polymerase chain reaction greatly facilitates generation of
mutants in vitro and the synthesis of
DNA for physical experiments.
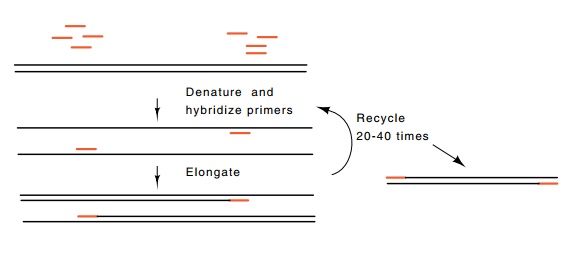
Figure
10.7 Polymerase chain reaction to
amplify the sequence containedbetween the sites to which two primers hybridize.
The polymerase chain reaction is a scheme that amplifies the DNA lying between two sequences which are within several thousand base pairs of one another. The amplification is
accomplished by first dena-turing the DNA sample, hybridizing two
oligonucleotide primers to the DNA, elongating with DNA polymerase, and
repeating this cycle up to 40 times (Fig. 10.7). The two oligonucleotide
primers must be comple-mentary to opposite strands of the DNA. The product of
elongation primed by one oligonucleotide plus the template can then become
templates for the next round of synthesis. As a result, each round of synthesis
doubles the number of product DNA molecules present. The first round of
synthesis produces DNA extending in one direction beyond each primer, but the
DNA made in subsequent cycles from the first product DNA extends just to the
ends of the primers.
Although DNA polymerase I from E. coli could be used in
the polymerase chain reaction, its use would be inefficient because each round
of denaturing the double-stranded DNA to form the single strands necessary as
templates would destroy the polymerase. Therefore, these procedures use a temperature-resistant
polymerase isolated from a thermophile, Thermus
aquaticus. This polymerase withstands the 95° incubation for denaturing the DNA. Even better, after an incubation at
45° to hybridize primer to the DNA, a temperature at
which the polym-erase is largely inactive, the temperature can be raised to 75° to activate the polymerase. Although a little of the primer dissociates
from the template at this temperature, a much greater fraction of any
incorrectly hybridized primer dissociates from incorrect sites. Thus, a very
great specificity is achieved for amplification of just the desired sequence of
DNA.
The polymerase chain reaction can be put to a wide
variety of uses. One simple example is the screening of cloning steps.
Ordinarily, one must screen transformants after the simple step of inserting a
DNA fragment into a plasmid. Typically 90% of the transformants contain the
fragment, but since the frequency is not 100%, one must verify that
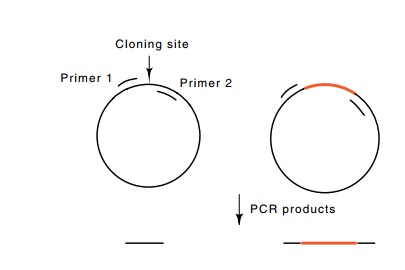
Figure
10.8 Polymerase chain reaction used to
monitor the insertion of aforeign DNA between the sites to which two primers
hybridize. Insertion of DNA between the primer hybridization sites increases
the size of the PCR product.
the clone selected for further work is suitable.
Previously such verifica-tion required growth of cultures from a collection of
candidates, puri-fication of the plasmids from each, and screening of each by
restriction enzyme digestion followed by gel electrophoresis to check for the
restriction fragment with properly altered size. PCR permits the same test in
much less time. Transformant colonies are picked directly into tubes used for
PCR. The first heat step lyses the cells. Primers are used that flank the site
into which the fragment was to be cloned. The product of the amplification is
run on a gel. If the fragment had been cloned between the sites, the amplified
piece will have one size, but if the fragment had not been cloned, the fragment
would be much smaller (Fig. 10.8).
DNA for footprinting or sequencing can be made
directly from genomic DNA with the polymerase chain reaction. By labeling one
of the oligonucleotide primers used in the PCR reaction, the DNA that is
synthesized is already radioactive and ready for use. Such a technique eliminates
the need for cloning the DNA from mutants. It also stream-lines the screening
of genetic diseases or of mutants isolated in the lab. Instead of cloning the
DNA to determine the defect in a gene, the DNA from the organism or cells can
be directly amplified and the defect determined by sequencing. PCR also greatly
facilitates genetic construc-tions. For example, suppose a portion of a gene is
to be cloned in an expression vector. The oligonucleotide primers can include
not only regions homologous to the DNA to be cloned, but also additional
regions necessary for cloning and expression like restriction sites, ribosome
binding sites, and translation termination signals (Fig. 10.9). In the first
round of amplification, the regions of homology between the primers and the
template DNA hybridize. In subsequent rounds, the entire
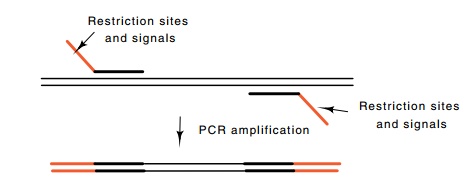
Figure
10.9 Use of PCR to both amplify a
selected region of DNA as well asplace specific desired sequences on the ends
of the DNA.
Related Topics