Chapter: Genetics and Molecular Biology: Advanced Genetic Engineering
Footprinting, Premodification and Missing Contact Probing
Footprinting, Premodification and Missing
Contact Probing
Understanding the biochemical mechanisms underlying
the regulation of gene expression is a central problem in biochemistry-biology.
One of the first steps in the study of a protein that binds to DNA is to
determine where it binds. The use of DNAse footprinting in conjunction with
Southern transfers to determine the locations of histone binding was briefly
described. Galas and Schmitz developed the elegant method of footprinting to
solve the problem of determining the location
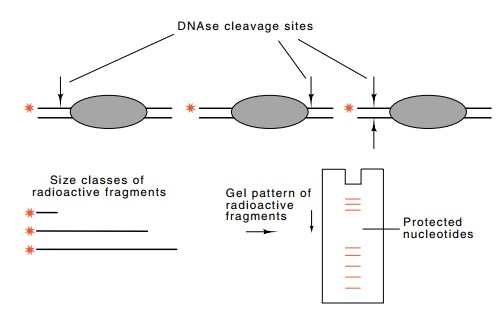
Figure
10.16 DNAse footprinting. Light
digestion by DNAse I nicks each of thelabeled DNA molecules on average one
time. No nicking occurs on any of the molecules under the bound protein and
therefore no fragments will be pro-duced with ends in this region.
at which a protein binds to DNA. As described
earlier, their method is based on the principles behind DNA sequencing.
Although it was de-vised for mapping DNAse-sensitive sites, it can be used with
any reagent that cleaves the DNA or modifies it so that later the DNA can be
cleaved. The basic idea can also be used in two modes, a protection mode in
which the bound protein protects the DNA from the reagent, and a prebinding
interference mode in which the DNA is modified first and then the positions of
modification which prevent protein binding are determined.
In the simplest form of footprinting the DNA
fragment to be investi-gated must first be labeled on just one end of one
strand. This can be done with polynucleotide kinase, with terminal transferase,
which adds nucleotides to the 3’-OH end of DNA, with DNA pol I, which fills out
sticky ends left by many restriction enzymes, or with PCR by amplifying a
region containing the binding site and the use of one radioactively-la-beled
oligonucleotide primer.
After the binding of a protein to the DNA, the
complex is briefly treated with DNAse. The duration of this treatment is
adjusted so that about one random strand scission occurs per DNA molecule.
Conse-quently, the population of molecules will contain examples of
phos-phodiester bond breakage at all positions except those covered by the
protein (Fig. 10.16). Then the DNA is denatured and subjected to
electrophoresis on a sequencing gel. This separates the population of molecules
according to size, and the amount of DNA in any band is proportional to the
amount of cleavage that occurred at the correspond
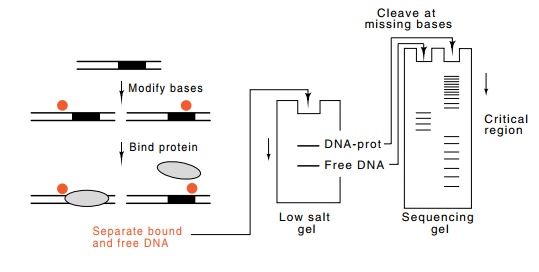
Figure
10.17 Premodification to locate
contacted bases or phosphates. Theintensity of the bands ultimately produced is
proportional to the numbers of DNA molecules that were modified at that
position. By binding protein to the population of DNA molecules, the subset
modified in regions essential for protein binding are separated from the subset
modified in irrelevant positions.
The footprinting experiments can also be performed
with the protein protecting the DNA binding site from chemical attack rather
than enzymatic attack. Dimethylsulfate can methylate guanine residues ex-cept
some of those protected by the protein. After the methylation, the DNA can be
cleaved at each of the methylated guanines, and the denatured, labeled
fragments can be subjected to electrophoresis on a sequencing gel. Both DNAse I
and dimethylsulfate are imperfect in that their reaction with unprotected DNA
is somewhat base-specific. Conse-quently, both of these techniques must be
performed with the control of labeled DNA free of the binding protein. The
differences in the intensities of the bands between the DNA and protein plus
DNA samples then identify the phosphodiester bonds protected from cleavage by
the protein. The hydroxyl radical will attack and cleave the phosphodiester
backbone independent of sequence. Thus, it is particularly useful for
footprinting experiments.
The premodification interference mode for
performing footprinting types of experiments is to modify or nick the DNA
before addition of the protein whose binding site is to be mapped (Fig. 10.17).
Then the protein is added. Those DNA molecules still capable of binding the
protein are separated from the DNA molecules that have been modified in regions
essential for binding of the protein, and as a result, do not bind the protein.
These two populations of DNA molecules are separated from one another by the
mobility retardation assay, in which DNA with a bound protein migrates more
slowly than DNA without a bound protein. If necessary, the two populations are
cleaved at the positions of modified
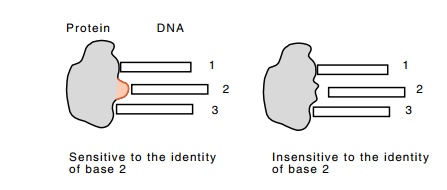
Figure
10.18 Changing a base-contacting amino
acid to an alanine can makethe protein insensitive to the identity of that
base.
The basic ideas of footprinting and premodification
probing can even be adapted to reveal specific amino acid residue-DNA base
interactions in the binding of a protein to DNA. The approach is similar to the
premodification method just described. The labeled DNA is treated chemically so
that each molecule, on average, has one base removed from a random position.
This can be either a hydroxyl radical treatment or one of the base-specific
reactions used in Maxam-Gilbert DNA sequencing. Then the entire population of
molecules is permitted to bind to the protein. There is no effect on the
binding if a base is missing from a position not contacted by the protein. If,
however, a base is missing from a position the protein contacts, the protein
will bind less tightly. Thus, if the protein and treated DNA are mixed and then
diluted so that no further binding can occur, the protein will dissociate
first, if it ever binds at all, from those DNA molecules that are missing bases
that are contacted by the protein. The population of free DNA molecules becomes
enriched with such DNA molecules. Similarly, the population of DNA molecules
with bound protein becomes enriched with molecules that are missing only those
bases not involved with contacts to the protein. The two DNA populations can be
separated from one another with the DNA band shift assay. If necessary, the
molecules are chemi-cally cleaved at the positions of missing bases, and the
positions of the cleavages are displayed after electrophoresis on DNA
sequencing gels.
To demonstrate a specific residue-base interaction,
the residue is modified by site-specific mutagenesis or PCR to an alanine.
Because alanine is smaller than most other amino acids, most likely it will be
unable to make the contact made by the amino acid it replaced (Fig. 10.18).
Thus, in performing the missing contact experiment, a new base will be in the
collection of bases not contacted. This is the base contacted by the amino acid
at the position of the new alanine. Of course, in the execution of the
experiment, allowance must be made for the fact that dissociation of the
protein from the DNA is faster because of the missing contact. Either a shorter
time is allowed for dissociation or buffer conditions are altered to increase
the affinity of binding.
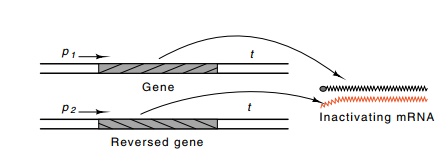
Figure
10.19 Generation of antisense RNA by
inverting a gene or a segmentof a gene and its subsequent hybridization in vivo to the mRNA from the gene,
thereby preventing its translation.
Related Topics