Chapter: Genetics and Molecular Biology: Advanced Genetic Engineering
Isolation of Rare Sequences Utilizing PCR
Isolation of Rare Sequences Utilizing PCR
The power of PCR can be used to simplify the
cloning of genes and to isolate special rare DNA or RNA molecules out of large
populations containing many different sequences. First let us consider the
cloning of a gene if a tiny portion of its protein product can be isolated. The
amino acid sequence of a stretch of the protein must be determined. This is
used to design oligonucleotides that hybridize to the top and bottom strands of
the ends of the region encoding the peptide and can be used in PCR to amplify
the region from cDNA template. Of course, redundancy in the genetic code
necessitates that each oligonucleotide be a mixture as discussed earlier. For
use with PCR, however, there is less need to minimize the degeneracy. Even
though the proper DNA sequence is represented by only a tiny fraction of each
oligonucleotide mixture, just these oligonucleotides will be functional in PCR
amplification of the desired region of DNA. The other oligonu - cleotides may
hybridize to the template cDNA, but they are unlikely to give rise to any PCR
products, and it is even less likely that such products would be of the same
size as the desired PCR product.
The PCR amplified product of a portion of the gene
can then be used to probe a cDNA library contained on plasmid or phage clones.
This will reveal the clones likely to contain an intact version of the desired
gene.
PCR is also useful for the isolation of very rare
sequences present in complex mixtures of sequences. During the chemical
synthesis of DNA, the use of mixtures of nucleotide precursors for some
synthesis cycles enables large random populations to be synthesized. Consider
the determination of the optimum binding sequence of a DNA-binding protein. One
strand of DNA can be synthesized with unique sequences at each end and
containing a totally random interior region. First, strands complementary to
the chemically synthesized molecules must be constructed. Due to the random
sequences involved, the complemen-
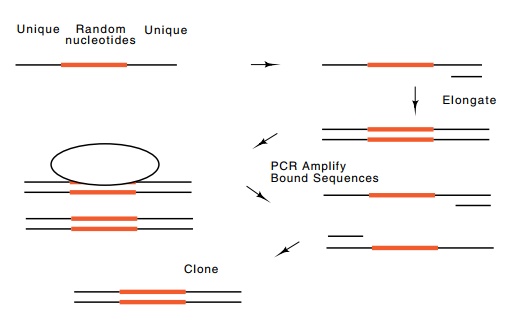
Figure
10.10 Use of PCR to amplify a small
amount of DNA present in acomplex mixture.
tary strands cannot be synthesized chemically. They
are made enzymati-cally be hybridizing a DNA primer to the unique sequence at
one end of the chemically synthesized strand. Next, DNA pol I is used to
elongate and complete the synthesis of an exact complement (Fig. 10.10). Then,
the small population of DNA molecules with sequences capable of binding the
protein are isolated, for example, by electrophoretic sepa-ration of
protein-DNA complexes from DNA incapable of binding the protein. This tiny
amount of DNA is amplified by PCR using primers complementary to the unique
sequences at the ends. After amplifica-tion, the selection and amplification
steps can be repeated, and finally the DNA can be cloned and sequenced.
Simple variations on the basic idea described above
can be developed for the isolation of RNA capable of binding a protein or
specific support on an exchange column. It is also possible to devise methods
for the selection of DNA or RNA molecules that possess hydrolytic activities.
Mutagenic steps can be introduced in the PCR
amplification steps so that variants of the selected molecules can be created.
In this way, true in vitro evolution
experiments can be done. Living cells are not required.
Related Topics