Chapter: Biotechnology Applying the Genetic Revolution: Bacterial Infections
Molecular Approaches to Diagnosis
MOLECULAR APPROACHES TO DIAGNOSIS
A major contribution of
molecular biology has been the development of improved methods for diagnosis.
In practice diagnosis usually means identifying the agent of disease,
whether a bacterium, virus, or protozoan. Some pathogenic bacteria grow slowly
or not at all when cultured outside their host organisms. Viruses are obligate
parasites and can only be grown in the laboratory by infecting appropriate
cultured host cells. Furthermore, different microorganisms require different
culture media and culture conditions. All these factors make traditional
methods of identification laborious. In contrast, molecular approaches analyze
macromolecules such as DNA, RNA, or protein rather than attempting to grow the disease
agents. Some new methods involve the use of antibody technology and are dealt .
Here we will consider nucleic acid–based approaches.
Molecular methods usually
start with extraction of DNA from either cultured pathogens or an infected
patient. The DNA is often amplified by PCR and then analyzed by sequencing or
hybridization. Thus the same reagents and procedures may be used for many
different microorganisms. Moreover, molecular methods are often quicker, more accurate,
and more sensitive than classical microbiological techniques. Most of the
molecular techniques used in clinical diagnosis have already been described
elsewhere in this book. Here we will consider their applications.
Every species of organism
has a different small-subunit ribosomal RNA sequence ( ssu rRNA ; 16S rRNA in
bacteria, 18S rRNA for eukaryotes). Hence bacteria and eukaryotic parasites may
be identified by analysis of their ssu rRNA sequences. In practice, the gene encoding
the ssu rRNA is sequenced, rather than the RNA itself.
(a) Ribotyping may be done by detailed
restriction analysis of the rRNA genes. DNA from a bacterial strain is digested
with several different restriction enzymes and the fragments are separated by
gel electrophoresis. Thefragments are transferred to a membrane and subjected to
Southern blotting to identify bands. A probe that corresponds to part of the
16S rRNA sequence is used. Many bacteria have more than one 16S rRNA gene and so
several bands will be seen for each digestion. This approach needs significant
amounts of DNA.
(b) Amplification of DNA by PCR followed by
DNA sequencing allows identification using only tiny amounts of DNA. Primers
that recognize the conserved region of 16S rRNA are used to generate a segment of
the 16S gene by PCR. The PCR fragment is then sequenced and compared with a
database of known DNA sequences. Note that the same procedure and reagents are
applicable to all bacterial infections. Furthermore, this method works with
bacteria that cannot be cultured. Variants of PCR, such as RAPD (randomly
amplified polymorphic DNA;), may also be used to identify the pathogen. For
bacterial RAPD-PCR, a random mixture of six-base primers is often used. This
allows different strains of the same bacterial species to be distinguished and
has been used to track the source and spread of contaminating bacteria in food
or water.
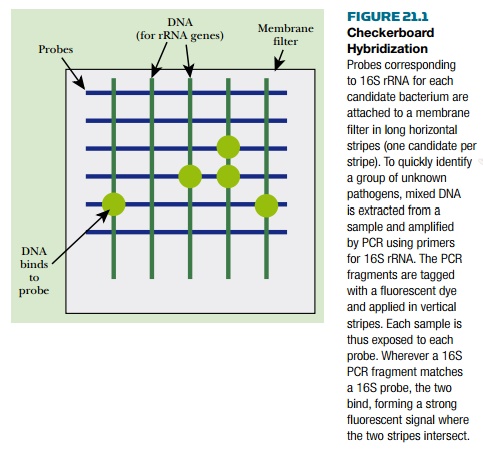
(c) Checkerboard hybridization is a technique
that allows multiple bacteria to be detected and identified simultaneously in a
single sample. A series of probes corresponding to different bacteria are
applied in horizontal lines across a hybridization membrane ( Fig. 21.1 ). PCR
is used to amplify a portion of the 16S gene from the target bacteria or clinical
samples, which may contain a mixture of bacteria. The PCR fragments are then
labeled with a fluorescent dye and applied vertically to the membrane. After
denaturation and annealing to allow hybridization, the membrane is washed to
remove unbound DNA. Those samples that hybridize to the probes appear as bright
fluorescent spots.
Related Topics