Chapter: Genetics and Molecular Biology: Repression and the lac Operon
Migration Retardation Assay and DNA Looping - Repression and the lac Operon
The Migration Retardation Assay and DNA Looping
Earlier it was mentioned that DNA can be subjected
to electrophoresis under conditions compatible with protein binding. For DNA
fragments in the size range of 50 to about 2,000 base pairs, the binding of a
protein significantly retards the migration. Therefore, free DNA and
protein-DNA complexes may easily be separated by electrophoresis and
identi-fied by staining or autoradiography. An additional virtue of the
migration retardation assay is the fact that protein and DNA can be incubated
in buffers containing physiological concentrations of salt, on the order of 50
mM KCl. Then, the electrophoresis can be performed at very low salt
concentration. As discussed earlier, salt has a dramatic effect on the affinity
with which most proteins bind to DNA. At low salt concentrations, most
proteins’ affinity is much higher than at physi-ological salt concentrations.
This greatly reduces the dissociation rate of the proteins. Additionally, the
presence of the gel surrounding the protein-DNA complex cages the protein and
further reduces its effective dissociation rate during electrophoresis. These
features make the gel migration retardation assay particularly useful for the
study of protein-DNA interactions as electrophoresis “freezes” a particular
solution condition.
Not surprisingly, the lac repressor-operator interaction can be stud-ied with the
migration retardation assay. The tetrameric lac
repressor can bind with two of its subunits to the main operator at the
promoter. Repressor’s other two subunits are free to bind to either of the
pseudo-operators that are located one hundred and four hundred base pairs to
either side. Such a double binding by a single repressor tetramer forms
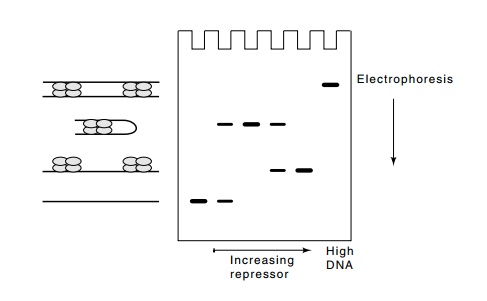
Figure
11.8 Migration retardation assay withlacrepressor and DNA contain-ing two
operators. As repressor concentration is increased, loops, then linear
structures are formed. At high DNA concentrations, sandwiches form.
a DNA loop. In
vivo this looping reaction is facilitated by supercoiling. The same looping
reaction can be facilitated in vitro
by using a linear DNA fragment containing two strong repressor-binding sites
separated by 100 base pairs. Incubation of such DNA with low concentrations of
repressor permits binding of single tetramers to DNA molecules. These form DNA
loops so that each tetramer contacts two operators from the same DNA molecule.
Incubation at higher repressor concentrations forces a separate repressor
tetramer onto each operator, and incubation at high DNA concentrations forms
structures in which repressors join two DNA molecules in a sandwich structure
(Fig. 11.8).
Related Topics