Chapter: Genetics and Molecular Biology: Repression and the lac Operon
DNA-binding Domain of lac Repressor
The DNA-binding Domain of lac Repressor
How could genetic experiments determine the amino acid residues that contact operator? Work by Miller and collaborators showed that non-sense mutations lying early in the lacI gene did not abolish expression of the entire gene. Although the ribosomes terminate translation on reaching the nonsense codon, often they do not dissociate from the mRNA before reinitiating translation at a site within the lacI gene. As a result, they synthesize a repressor molecule that lacks up to 61 of the amino acids normally found at the N-terminus. Most surprisingly, these truncated repressor polypeptides fold, associate as usual to form tetra-mers, and bind IPTG. They are, however, incapable of binding to DNA as shown in vivo by their inability to repress and in vitro by the DNA filter-binding assay. The simplest explanation of their inability to bind to DNA is that the amino acids missing from their N-terminus are the part of the wild-type repressor that recognizes and binds to operator. In addition, more detailed genetic analysis of the repressor have shown that the overwhelming majority of missense mutations affecting DNA binding by the repressor map in the region of the I gene coding for the first 60 amino acids.
A typical
LacI- mutant is recessive to the wild-type lacI gene. The amino-terminal truncated repressors, however, are
dominant negatives. That is, the diploid lacI+/lacI-d behaves like an I-.
As explained, the dominance of I-d
mutations to the I+ allele
results from the tetrameric structure of repressor and the fact that all four
subunits are required for full repression.
The steps
to combine to isolate a particular mutant sometimes are not apparent, so we
will list and explain a series of genetics steps designed to isolate an I-d
mutant.
1.
Isolate I- mutations
using phenyl-β-D-galactoside
plates. Since phenyl-gal is not an inducer, but is a substrate of β-galactosidase, any cells growing
on these plates must be constitutive.
2.
Score to determine which are
nonsense I- mutations. The muta-tions can be isolated on an episome,
F’lac+pro+. To
test, the episome can be mated into an F- su+ strain.
This strain should be deleted of lac
and pro genes. To determine whether
the episome in the suppressing strain
is I+
or I-, the plates can include 5-bromo-4-chloro-3-indolyl-β−D-D-galactoside (X-gal). If X-gal
is included in minimal glycerol plates, the constitutives will form deep blue
colonies because the hydrolysis of this substrate by β-galactosidase produces an
insoluble blue dye.
3. Map
the locations of the I- mutations. This can be done using the
ability of strains containing the mutation on an episome and a deletion into
the I gene on the chromosome to reconstruct an I+ gene (Fig. 11.11).
A point mutation is able to recombine with deletion 1 but not deletion 2 to
form I+ recombinants. Although this type of mapping only positions
the mutation to the left or right of the end point of the deletion, an ordered
set of deletions can be used to locate the mutation to within about 10 base
pairs.
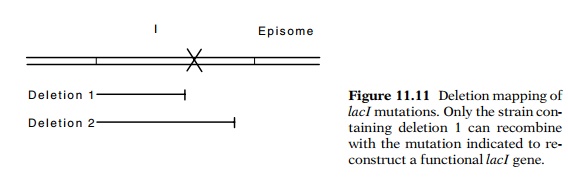
How can the selective plates be arranged to permit growth of only the cells that can reconstruct a functional I gene? The plates must prevent growth of each of the parent types of cells as well as mated cells that are unable to form a wild-type lacI gene. This can be done by using phenyl-gal and glycerol in plates. The deletion mapping can be done in a GalE- strain. The resulting defective galactose epimerase gene renders the cells sensitive to galactose. The LacI- cells are constitutive and cleave the phenyl-gal to produce galactose, which then prevents their growth. A LacI+ recombinant, however, represses β-galactosidase synthesis and does not generate galactose. Such cells can continue to grow in the presence of phenyl-gal.
4.
Test the candidates for being trans-dominant repressor negative. This
is done by mating the episome into an I+ strain and selecting for
transfer of the pro marker. This
strain is tested by spotting onto X-gal plates as before.
5.
Physical studies of the repressor
from the I-d mutants can test whether the repressor is a tetramer,
and SDS gels can show that the mutated repressor is shorter than wild-type.
Protein
sequencing can provide the final proof that the repressor synthesized in the
nonsense I-d mutants results from translational restarts. The mutant
repressor can be purified by precipitation with antibody, separated from the
antibody by SDS gel electrophoresis, eluted from the gel, and the N-terminal
amino acids sequenced.
Related Topics