Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Influenza, Respiratory Syncytial Virus, Adenovirus, and Other Respiratory Viruses
Influenza A
Influenza A
Influenza A is considered in detail because of its great clinical and epidemiologic importance.
The influenza A virion contains eight segments of single-stranded RNA with defined genetic responsibilities. These functions include coding for virus-specified proteins (see Fig 33–1; Table 33–2). A unique aspect of influenza A viruses is their ability to develop a wide variety of subtypes through the processes of mutation and whole-gene “swapping” between strains, called reassortment.Recombination, which occurs when new genes are assembled from sections of other genes, is thought to occur rarely, if at all. These processes result in antigenic changes called drifts and shifts, which are discussed shortly.
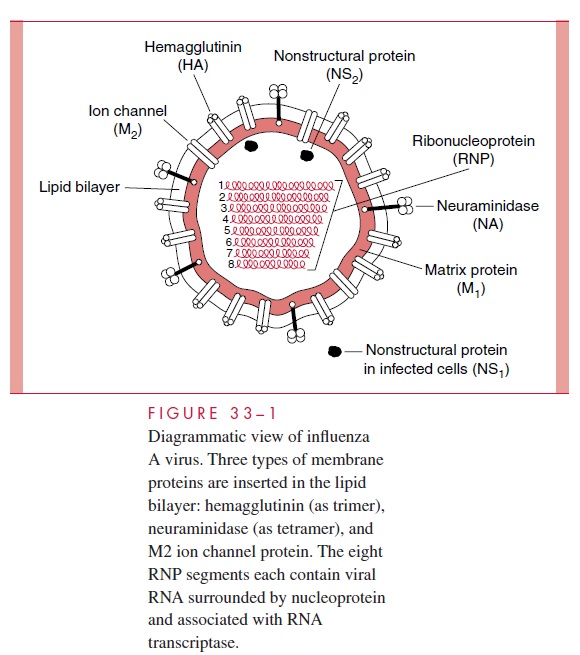
The 15 recognized subtypes of hemagglutinin and 9 neuraminidase subtypes known to exist among influenza A viruses that circulate in birds and mammals represent a reservoir of viral genes that can undergo reassortment, or “mixing” with human strains. Three hemagglutinins (H1, H2, and H3) and two neuraminidases (N1 and N2) appear to be of greatest importance in human infections. These subtypes are designated according to the H and N antigens on their surface (eg, H1N1, H3N2). There may also be more subtle, but sometimes important, antigenic differences (drifts) within each subtype. These differences are designated according to the major representative virus to which they are most closely related antigenically, using the place of initial isolation, number of the isolate, and
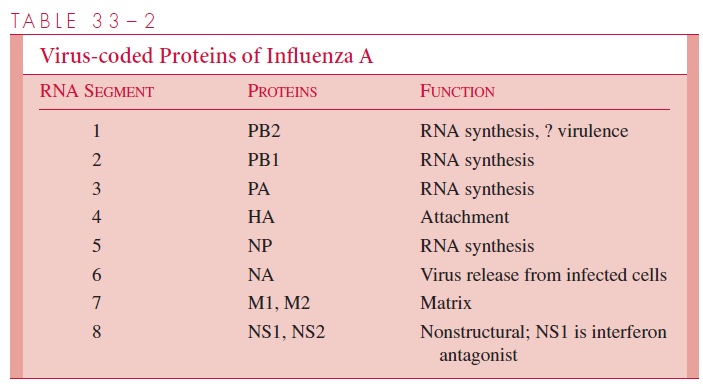
Antigenic drifts within major subtypes can involve either H or N antigens, as well as the genes encoding nonstructural proteins, and may result from as little as a single muta-tion in the viral RNA. The mutant may come to predominate under selective immuno-logic pressures in the host population. Such drifts are frequent among influenza A viruses, occurring at least every few years and sometimes even during the course of a single epidemic. Drifts can also develop in influenza B viruses but at a considerably lower frequency.
In contrast to the frequently occurring mutations that cause antigenic drift among influenza A strains, major changes (>50%) in the nucleotide sequences of the H or N genes can occur suddenly and unpredictably. These are referred to as antigenic shifts. They almost certainly result from reassortment that can be readily reproduced in the lab-oratory. Simultaneously infecting a cell with two different influenza A subtypes yields progeny that contain antigens derived from either of the original viruses. For example, a cell infected simultaneously with influenza A (H3N2) and influenza A (H1N1) may produce a mixture of influenza viruses of the subtypes H3N2, H1N1, H1N2, and H3N1. When novel “new” epidemic strains emerge, they most likely have circulated into animal or avian reservoirs where they have undergone genetic reassortment (and sometimes also mutation), then readapted and spread to human hosts when a sufficient proportion of the population has little or no immunity to the “new” subtypes. A recent example was the appearance in Hong Kong in 1997 of human cases caused by an avian influenza A (H5N1). Studies indicated that all RNA segments were derived from an avian influenzaA virus, but a single insert coding for several additional amino acids in the hemagglu-tinin protein facilitated cleavage by human cellular enzymes. In addition, a single amino acid substitution in the PB2 polymerase protein occurred. These two mutations together made the virus more virulent for humans; fortunately, human-to-human transmission was poor.
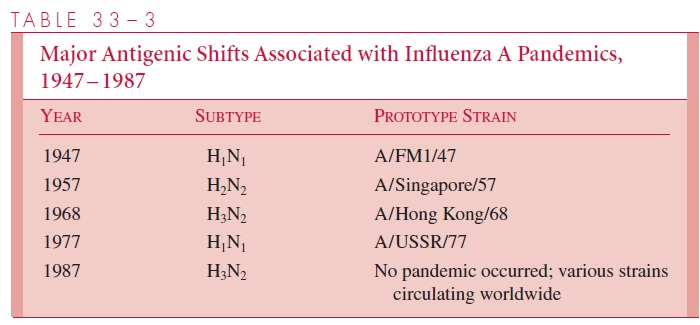
Major antigenic shifts, which occurred approximately every 8 to 10 years in the 20th century, often resulted in serious epidemics or pandemics among populations with littleor no preexisting antibody to the new subtypes. Examples include the appearance of an H1N1 subtype in 1947, followed by an abrupt shift to an H2N2 strain in 1957, which caused the pandemic of Asian flu. A subsequent major shift in 1968 to an H3N2 subtype (the Hong Kong flu) led to another, but somewhat less severe epidemic. The Russian flu, which appeared in late 1977, was caused by an H1N1 subtype very similar to that which dominated between 1947 and 1957 (Table 33–3).
The concepts of antigenic shift and drift in human influenza A virus infections can be approximately summarized as follows. Periodic shifts in the major antigenic components appear, usually resulting in major epidemics in populations with little or no immunologic
As the population of susceptible individuals is exhausted (ie, subtype-specific immunity is acquired by increasing numbers of people), the subtype continues to circulate for a time, undergoing mutations with subtle antigenic drifts from season to season. This allows some degree of virus transmission to continue. Infectivity persists because subtype-specific immunity is not entirely protective against drifting strains; for example, an individual may have antibodies reasonably protective against influenza A/Texas/77(H3N2), yet be susceptible in succeeding years to reinfection by influenza A/Bangkok/79(H3N2). Eventually, however, the overall immunity of the population becomes sufficient to minimize the epidemic potential of the major subtype and its drifting strains. Unfortunately, the battle is never entirely won; the scene is set for the sudden and usually unpredictable appearance of an entirely new subtype that may not have circulated among humans for 20 years or more.
Related Topics