Chapter: Biotechnology: Recombinant DNA Technology
Hybridisation Techniques - Recombinant DNA Technology
Hybridisation Techniques
Once a specific DNA sequence has been isolated by cloning, it can be used as a probe to detect the presence and amount of complementary DNA sequences present in isolated DNA, for example from different species. A question can be asked such as, Is this particular gene or DNA fragment present in a mammalian cell also present in an insect cell?
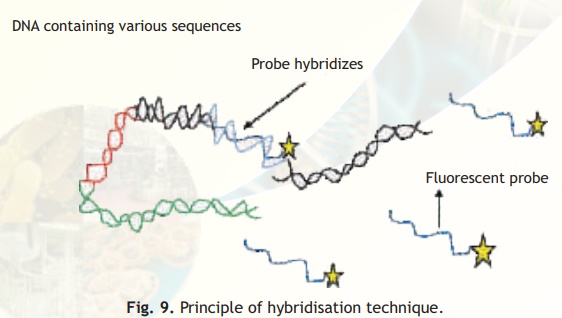
DNA probes are relatively small single stranded sequences of DNA that recognise and bind to complementary sequences. Recall that two strands of DNA are held together by base complementarity i.e., base A on one DNA strand hydrogen bonds with base T on the complementary strand and likewise base C hydrogen bonds with base G. A DNA probe which is single stranded will bind to a complementary sequence with the same base pairing rules (hybridisation) and if the probe can be tagged with a flourescent or radioactive label the complementary sequence can be located either in a cell nucleus or on a gel chromatogram. This principle is the basis of all hybridisation techniques, see Fig. 9.
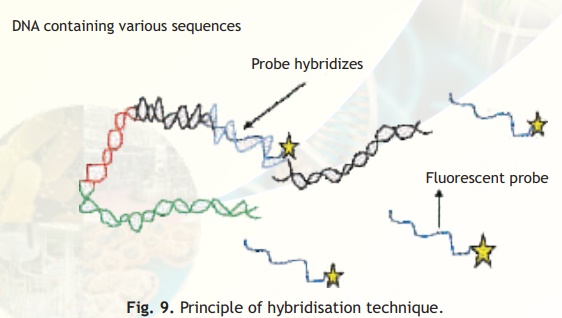
Fig. 9. Principle of hybridisation technique.
If the probe is tagged with a fluorescent label, under UV light its location can be easily seen as it will fluoresce. On the other hand if the probe is tagged with a radioactive label a technique called autoradiography is used wherein the gel is placed on a photographic film and the probe location is indicated by white spots on the developed film.
Southern Hybridisation Technique
This technique of identifying and locating specific sequences in DNA gels using probes was invented in 1975 by Edward Southern and is named Southern Hybridisation technique in his honor. This is an essential technique in all rDNA experiments and its aim is to identify a specific DNA sequence in a heterogenous population of DNA molecules. As discussed in the previous section the principle of the technique is based on the ability of a probe to seek out and bind to its complementary sequence.
The procedure involves isolation and digestion of total genomic DNA with one or more restriction enzymes. The DNA fragments thus generated are separated in agarose gels using the technique of electrophoresis. Following separation of the DNA fragments due to size differences, they are transferred from the gel to a nylon or nitrocellulose membrane in a technique called blotting, see Fig. 10. In the blotting procedure the DNA fragments are forced from the gel onto the membrane by capillary action. The membranes are baked briefly to fix the DNA fragments to the membrane so that they do not diffuse during the next step of hybridisation. Note that the membrane will have the same pattern of DNA bands as the original agarose gel. The membrane is then treated with the single stranded labelled probe for an appropriate period after which the membrane is washed and either photographed under UV light (if probe label is fluorescent) or overlaid with a photographic film (if probe is radioactive). The location of the probe is determined leading to the identification of a gene or specific DNA fragment obtained from that given genomic DNA. A similar principle is used for determining RNA locations and this technique is called Northern hybridisation.
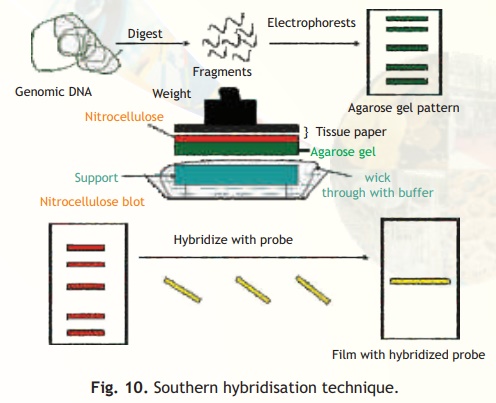
Fig. 10. Southern hybridisation technique.
Related Topics