Chapter: Biotechnology: Recombinant DNA Technology
DNA Sequencing
DNA Sequencing
In this era of genomics wherein whole genomes of species are being sequenced and compared to get a vision into the fundamental nature of DNA, the blueprint of life, the ease with which DNA is sequenced has played a major role. In the seventies two major methods were developed to sequence DNA:
1. The dideoxynucleotide chain termination method invented by Fred Sanger (the same scientist who invented a protein sequencing ) and Andrew Coulson
2. Chemical degradation method invented by Walter Gilbert and Allan Maxam (also called the Maxam and Gilbert method).
Of the two methods the first method is more popularily used and hence this technique will be discussed.
Dideoxynucleotide Chain Termination method
DNA polymerases, the major enzymes required in replication, have certain properties:
1. A single stranded DNA template is required for them to act upon.
2. A new strand cannot be initiated; only primers can be extended using the single strand DNA template as a guide.
3. Extension or DNA synthesis occurs in a 5'->3'direction which requires that a new nucleotide is added to the 3' hydroxyl group of the chain.
4. Deoxynucleotide 5' triphosphates are the normal substrates.
5. However if the 3' hydroxyl group of a deoxynucleotide triphosphate is absent as in the 2',3' dideoxynucleotide triphosphate (Fig. 12) it would result in, that nucleotide being incorporated into the growing chain but subsequently the chain cannot further be extended (no 3' hydroxyl) and this is the main principle of the Sanger Dideoxynucleotide Chain Termination method.
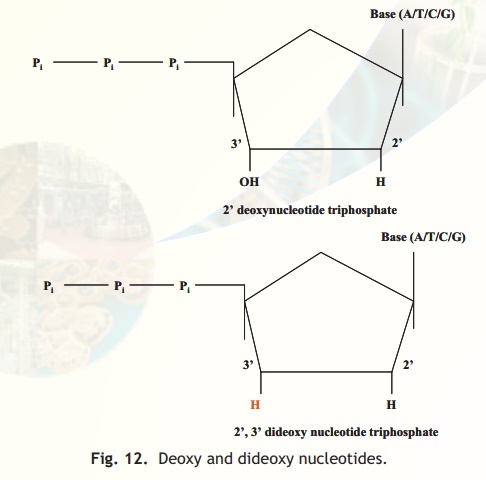
Fig. 12. Deoxy and dideoxy nucleotides
During a primer extension process, whenever a dideoxynucleotide is incorporated, the DNA polymerase enzyme cannot further carry out its reaction and the new DNA strand is thereby terminated (Fig. 13)
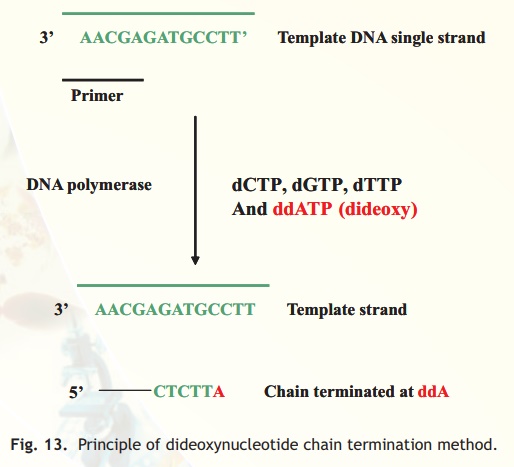
Fig. 13. Principle of dideoxynucleotide chain termination method.
Typically the sequencing technique is carried out in four test tubes. Each test tube carries the following common reagents, single stranded DNA template, deoxynucleotide triphosphates, primer and DNA polymerase. In addition to it a small amount of the four dideoxynucleotide triphosphates i.e ddATP, ddCTP, ddGTP and ddTTP is added separately into the four test tubes. The purpose of adding only a small amount of the dideoxy derivative is that only one dideoxy derivative is incorporated into each extending chain causing termination. Hence in a test tube containing ddATP all chains will end at ddA but at different positions of T present in the template (Fig. 14). The same is applicable to the other test tubes containing ddCTP, ddGTP and ddTTP. It is to be noted that the dideoxynucleotide is added wherever the complementary base is present on the template.
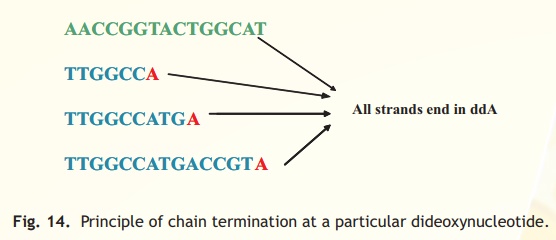
Fig. 14. Principle of chain termination at a particular dideoxynucleotide.
The various strands prematurely terminated at the particular ddNTP in a given tube are subjected to electrophoresis in special gels wherein bands can be resolved (separated) even if they differ by one nucleotide. The strands migrate in the gel with the shorter fragments moving faster towards the anode. The primers used in each tube can be made radioactive and hence the position of the separated strands in the gel chromatogram can be easily visualised using autoradiography. Fig. 15 is a schematic representation of a typical gel and its read sequence. Colours for different dideoxy nucleotides are for clarity.
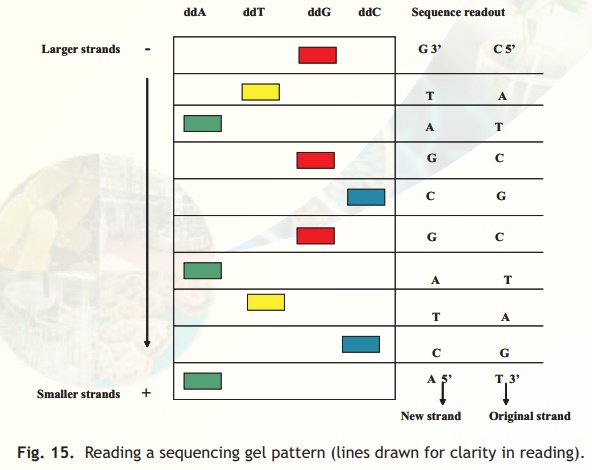
Fig. 15. Reading a sequencing gel pattern (lines drawn for clarity in reading).
Nowadays DNA sequencing technologies have become automated. To avoid using radioisotopes and their consequent danger, dideoxynucleotides are conjugated with florescent molecules which on excitation give a different colour each. Hence each band on the gel (read from anode to cathode) indicates the particular base as its terminal dideoxy nucleotide fluoresces with a given colour. This avoids the use of a four lane gel, a single lane gel electrophoresis is instead conducted and the gels are then laser scanned and the data fed into a computer. The computer is programmed to display the gel scan and the base readout as shown in Fig. 16.
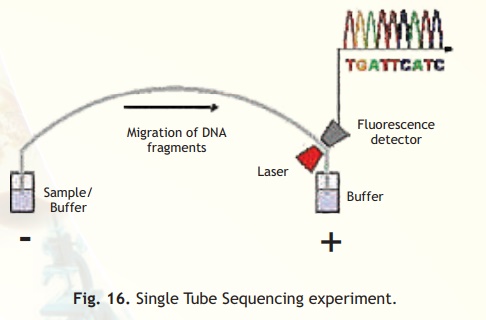
Fig. 16. Single Tube Sequencing experiment.
Related Topics