Chapter: Essential Microbiology: Viruses
Viral replication cycles
Viral replication cycles
One characteristic viruses share in common with true living organisms is the need to reproduce themselves∗ . As we have seen, all viruses are obligate intracellular parasites, and so in order to replicate, a host cell must be successfully entered. It is the host cell
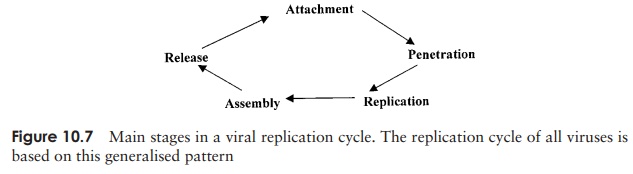
that provides much of the ‘machinery’ necessary for viral replication. All viral growth cycles follow the same general sequence of events (Figure 10.7), with some differences from one type to another, determined by viral structure and the nature of the host cell.
Replication cycles in bacteriophages
Viruses that infect bacterial cells are called bacteriophages (phages for short), which means, literally, ‘bacteria eaters’. Perhaps the best understood of all viral replication cycles are those of a class of bacteriophages which infect E. coli, known as the T-evenphages. These are large, complex viruses, with a characteristic head and tail structure(Figure 10.8). The double-stranded, linear DNA genome contains over 100 genes, and is contained within the icosahedral head. The growth cycle is said to belytic, because itculminates in the lysis (=bursting) of the host cell. Figure 10.9 shows the lytic cycle of phage T4, and the main stages are described below.
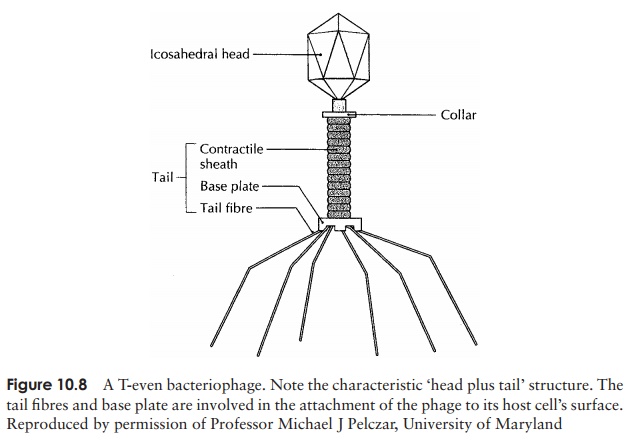
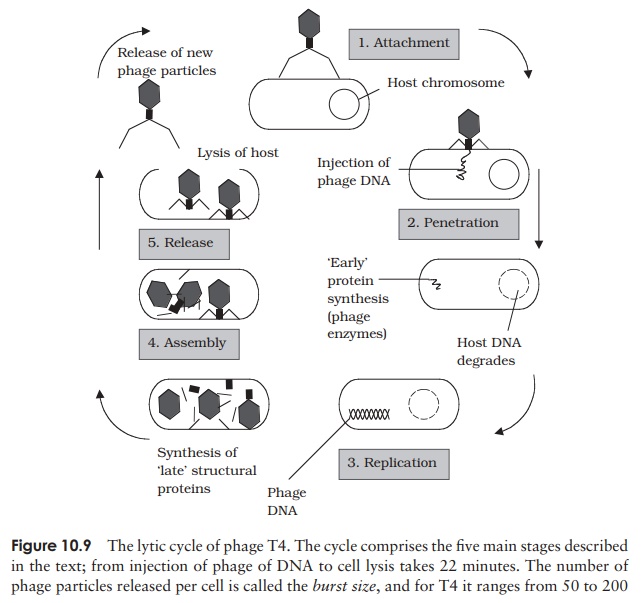
1. Adsorption (attachment): T4 attaches by means ofspecific tail fibre proteins to complementary recep-tors on the host cell’s surface. The nature of these receptors is one of the main factors in determining a virus’s host specificity.
2. Penetration: The enzyme lysozyme, present in thetail of the phage, weakens the cell wall at the point of attachment, and a contraction of the tail sheath of the phage causes the core to be pushed down into the cell, releasing the viral DNA into the in-terior of the bacterium. The capsid remains en-tirely outside the cell, as elegantly demonstrated in the famous experiment by Hershey and Chase.
3. Replication: Phage genes cause host protein and nu-cleic acid synthesis to be switched off, so that all of the host’s metabolic machinery becomes dedi-cated to the synthesis of phage DNA and proteins. Host nucleic acids are degraded by phage-encoded enzymes, thereby providing a supply of nucleotide building blocks. Host enzymes are employed to repli-cate phage DNA, which is then transcribed into mRNA and translated into protein.
3. Assembly: Once synthesised in sufficient quanti-ties, capsid and DNA components assemble sponta-neously into viral particles. The head and tail regions are synthesised separately, then the head is filled with the DNA genome, and joined onto the tail.
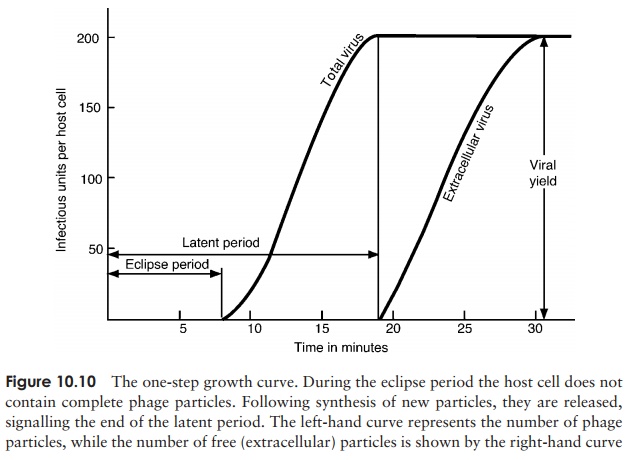
4. Release: Phage-encoded lysozyme weakens the cell wall, and leads to lysis of the celland release of viral particles;these are able to infect new host cells, and in so doing recommence the cycle. During the early phase of infection, the host cell contains components of phage, but no complete particles. This period is known as theeclipseperiod. The time which elapses between the attachment of a phage particle to the cellsurface and the release of newly-synthesised phages is the latent period(sometimes known as the burst time); for T4 under optimal conditions, this is around 22 minutes. This can be seen in a one-step growth curve, as shown in Figure 10.10.
Lysogenic replication cycle
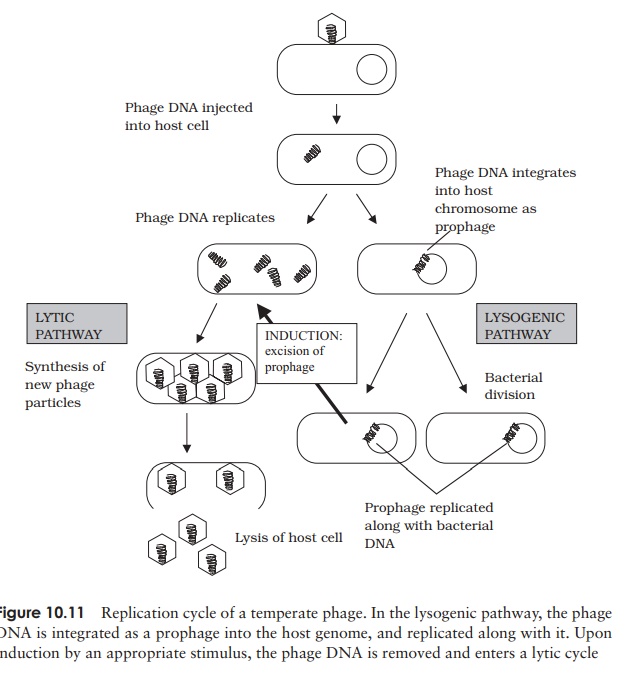
Phages such as T4, which cause the lysis of their cells, are termed virulent phages. Temperate phages, in addition to following a lytic cycle as outlined above, are able toundergo an alternative form of growth cycle. Here, the phage DNA actually becomes incorporated into the host’s genome as a prophage (Figure 10.11). In this condition of lysogeny, the host cell suffers no harm. This is because the action of repressor proteins,encoded by the phage, prevents most of the other phage genes being transcribed. These genes are, however, replicated along with the bacterial chromosome, so all the bacterial offspring contain the incorporated prophage. The lysogenic state is ended when the survival of the host cell is threatened, usually by an environmental factor such as UV light or a chemical mutagen. Inactivation of the repressor protein allows the phage DNA to be excised, and adopt a circular form in the cytoplasm. In this form, it initiates a lytic cycle, resulting in destruction of the host cell. An example of a temperate phage is bacteriophage λ (Lambda), which infects certain strains of E. coli. Bacterial strains that can incorporate phage DNA in this way are termed lysogens.
Replication cycles in animal viruses
Viruses that infect multicellular organisms such as animals may be specific not only to a particular organism, but also to a particular cell or tissue type. This is known as thetissue tropism of the virus, and is due to the fact that attachment occurs via specificreceptors on the host cell surface.
The growth cycles of animal viruses have the same main stages as described for bacteriophages (Figure 10.7), but may differ a good deal in some of the details. Most of these variations are a reflection of differences in structure between bacterial and animal host cells.
Adsorption and penetration
Animal viruses do not have the head and tail structure of phages, so it follows that their method of attachment is different. The specific interaction with a host receptor is made via some component of the capsid, or, in the case of enveloped viruses, by special structures such as spikes (peplomers). Viral attachment sites can frequently be blocked by host antibody molecules; however some viruses (e.g. the rhinoviruses) have overcome this by having their sites situated in deep depressions, inaccessible to the antibodies.
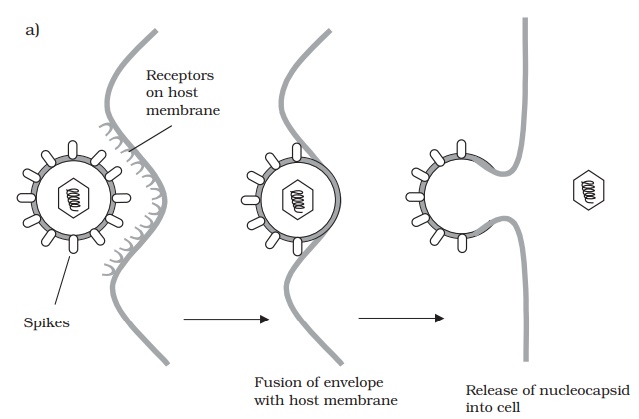
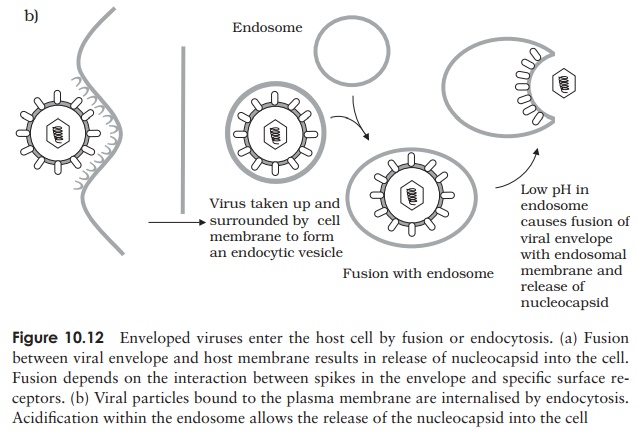
Whereas bacteriophages inject their nucleic acid component from the outside, the process in animal viruses is more complex, a fact reflected in the time taken for com-pletion of the process. Animal viruses do not have to cope with a thick cell wall, and in many such cases the entire virionis internalised. This necessitates the extra step of uncoating, a process carried out by host enzymes. Many animal viruses possess an en-velope; such viruses are taken into the cell either by fusion with the cell membrane, or by endocytosis (Figure 10.12). While some non-enveloped types release only their nu-cleic acid component into the cytoplasm, others require additionally that virus-encoded enzymes be introduced to ensure successful replication.
Replication (DNA viruses)
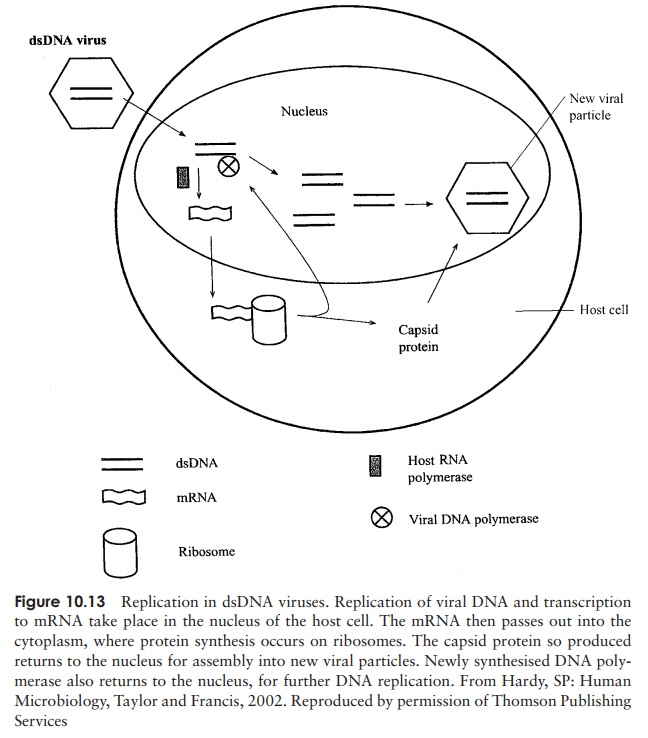
The DNA of animal cells, unlike that of bacteria, is compartmentalised within a nucleus, and it is here that replication and transcription of viral DNA generally occur∗ . Messenger RNA then passes to ribosomes in the cytoplasm for translation (Figure 10.13). In the case of viruses with a ssDNA genome, a double-stranded intermediate is formed, which serves as a template for mRNA synthesis.
Assembly
Translation products are finally returned to the nucleus for assembly into new virus particles.
Release
Naked (non-enveloped) viruses are generally released by lysis of the host cell. In the case of enveloped forms, release is more gradual. The host’s plasma membrane is modified by the insertion of virus-encoded proteins, before engulfing the virus particle and releasing it by a process of budding. This can be seen as essentially the reverse of the process of internalisation by fusion (Figure 10.12a).
Replication of RNA viruses
The phage and animal virus growth cycles we have described so far have all involved double-stranded DNA genomes. As you will remember, many viruses contain RNA instead of DNA as their genetic material, and we now need to consider briefly how these viruses complete their replication cycles.
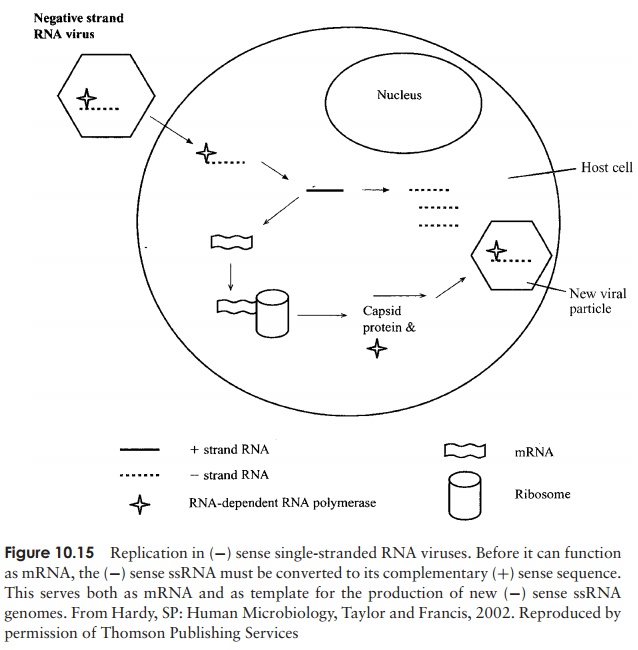
Replication of RNA viruses occurs in the cytoplasm of the host; depending on whether the RNA is single- or double-stranded, and (+) or (−) sense, the details differ. The genome of a (+) sense single stranded RNA virus functions directly as an mRNA molecule, producing a giantpolyprotein, which is then cleaved into the various struc-tural and functional proteins of the virus. In order for the (+) sense RNA to be replicated, a complementary (−) sense strand must be made, which acts as a template for the pro-duction of more (+) sense RNA (Figure 10.14). The RNA of a (−) sense RNA virus must first act as a template for the formation of its complementary sequence by a virally encoded RNA polymerase. The (+) sense RNA so formed has two functions: (i) to act
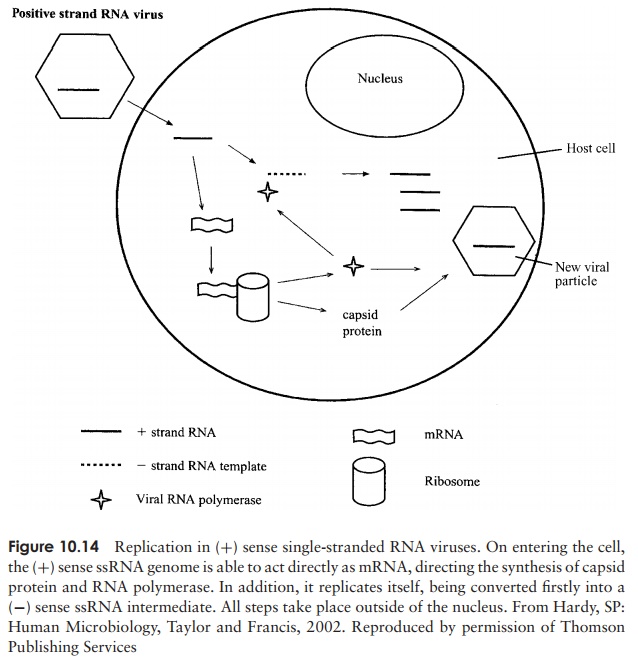
as mRNA and undergo translation into the virus’s various proteins, and (ii) to act as template for the production of more genomic (−) sense RNA (Figure 10.15).
Double-stranded RNA viruses are all segmented. They form separate mRNAs foreach of their proteins by transcription of the (−) strand of their genome. These are each translated, and later form an aggregate (subviral particle) with specific proteins, where they act as templates for the synthesis of a double-stranded RNA genome, ready for incorporation into a new viral particle.
Two final, rather complicated, variations on the viral replication cycles involve the enzyme reverse transcriptase, first discovered in 1970.
Retroviruses
These viruses, which include some important human pathogens, have a genome that exists as RNA and DNA at different part of their replication cycle. Retroviruses have a (+) sense ss-RNA genome which is unique among viruses in being diploid. The two copies of the genome serve as templates for the enzyme reverse transcriptase to produce
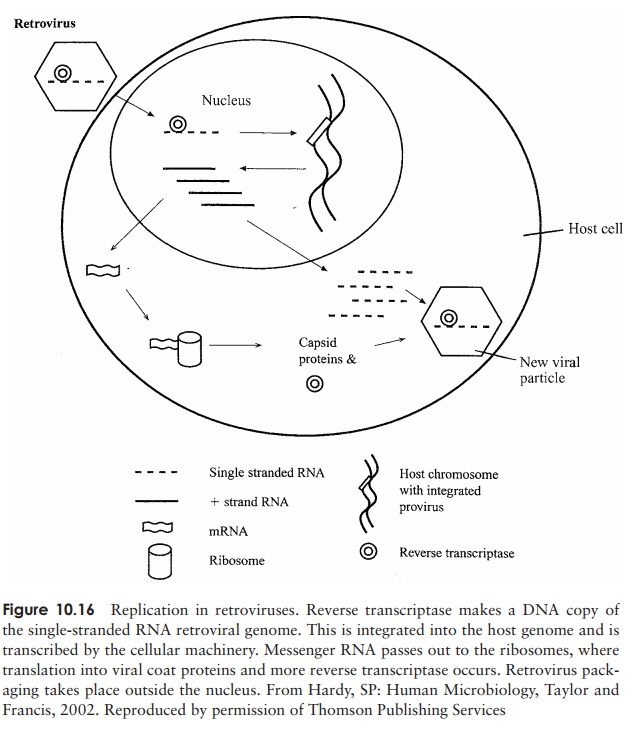
a complementary strand of DNA. The RNA component of this hybrid is then degraded, allowing the synthesis of a second strand of DNA. This proviral DNA passes to the nucleus, where it is incorporated into the host’s genome (Figure 10.16). Transcription by means of a host RNA polymerase results in mRNA, which is translated into viral proteins and also serves as genomic material for the new retrovirus particles. The human immunod-eficiency virus (HIV), the causative agent of acquired immune deficiency syndrome, is an important example of a retrovirus.
Hepadnaviruses
In two families of viruses (hepadnaviruses and caulimoviruses), DNA and RNA phases again alternate, but their order of appearance is reversed, so that a dsDNA genome is produced. This is made possible by reverse transcriptase occurring at a later stage, during the maturation of the virus particle.
Replication cycles in plant viruses
Viral infections of plants can be spread by one of two principal pathways. Horizontaltransmission is the introduction of a virus from the outside, and typically involves insectvectors, which use their mouthparts to penetrate the cell wall and introduce the virus. This form of transmission can also occur by means of inanimate objects such as garden tools. In vertical transmission, the virus is passed from a plant to its offspring, either by asexual propagation or through infected seeds.
The majority of plant viruses discovered so far have an RNA genome, although DNA forms such as the caulimoviruses are also known. Replication is similar to that of animal viruses, depending on the nature of the viral genome. An infection only becomes significant if it spreads throughout the plant (a systemic infection). Viral
Related Topics