Microbial Genetics - Types of Mutations | 12th Microbiology : Chapter 12 : Microbial Genetics
Chapter: 12th Microbiology : Chapter 12 : Microbial Genetics
Types of Mutations
Types of Mutations
The base sequence of DNA determines the amino acid sequence of a protein. The chemical and physical properties of each protein are determined by its amino acid sequence, so a single amino acid change is capable of altering the activity of, or even completely inactivating, a protein. Genotype refers to the genetic composition of an organism. Phenotype is an observable property of organism. The functional form of a gene is called Wildtype because presumably this is the form found in nature.
Mutation is the process by which the sequence of base pairs in a DNA molecule is altered.The alteration
can be a single base pair substitution, insertion or deletion.
Mutations can be divided into two general categories:
1. Base–pair substitution mutation involves
a change in the DNA such that one base pair is replaced by another.
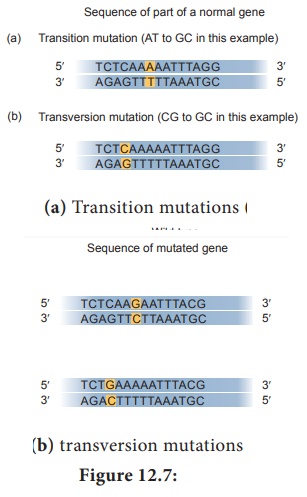
• A mutation from one purine – pyrimidine base pair to the other purine
–pyrimidine base pair is a transition mutation (Figure 12.7a). E.g. AT to GC,
CG to TA.
• A mutation from a purine - pyrimdine base pair to a pyrimidine purine
base pair is a transversion mutation (Figure 12.7b). E.g. AT to TA, CG to GC.
2. Base pair insertion or deletions involves the addition or deletion of one base pair.
If one or more base pairs are added to or deleted from a protein coding gene,
the reading frame of an mRNA can change downstream of the mutation. An addition
or deletion of one base pair, for example, shifts the mRNA’s downstream reading
frame by one base, so that incorrect amino acids are added to the polypeptide
chain after the mutation site. This type of mutation, called a frame shift
mutation (Figure 12.8) usually results in a nonfunctional protein.
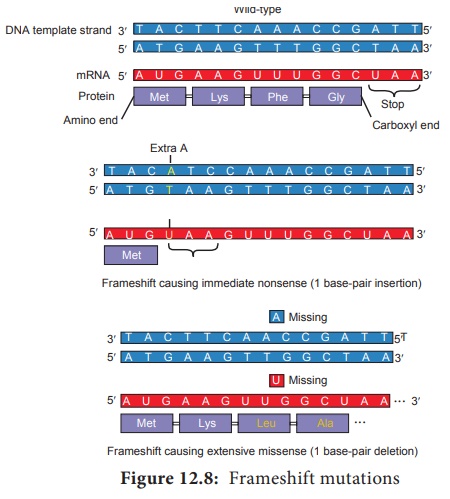
Frame shift mutations
• May
generate new stop codons, resulting in a shortened protein.
• May
result in a read through of the normal stop codon, resulting in longer than
normal proteins
• Or may result in a complete alteration of the amino acid sequence of a protein.
Point mutations are single base changes, that do not affect the
reading frame, that is, the mutation only makes a single change in a single
codon, and everything else is undisturbed.
Mutations can also be defined according to their
effects on amino acid sequences in proteins. They are
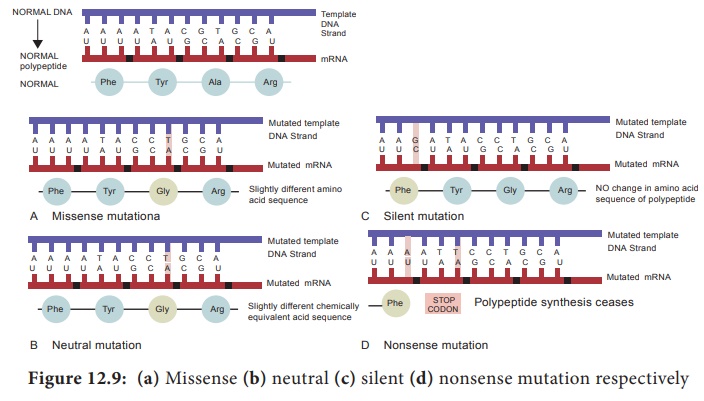
1. A missense mutation (Figure 12.9a) is a gene
mutation in which a base – pair change in the DNA changes a codon in an mRNA so
that a different amino acid is inserted into the polypeptide.
2. A
neutral mutation (Figure 12.9b) is a subset of missense mutations in which the
new codon codes for a different amino acid that is chemically equivalent to the
original and therefore does not affect the proteins function. Consequently, the
phenotype does not change.
3. A silent mutation (Figure 12.9c) is also a
subset of missense mutations that occurs when a base – pair change in a gene
alters a codon in the mRNA such that the same amino acid is inserted in the
protein. In this case, the protein obviously has a wild type function.
4. A
nonsense mutation (Figure 12.9d) is a gene mutation in which a base – pair
change in the DNA, changes a codon in an mRNA to a stop (nonsense) codon (UAG,
UAA or UGA). Nonsense mutation cause premature chain termination so instead of
complete polypeptides, shorter than normal polypeptide fragments (often
nonfunctional) are formed.
Forward mutations change the genotype from wild type to mutant and reverse mutations (or reversions or back mutations) change the genotype from mutant to wild type or to partially wild type. An organism which has reverted is a Revertant. The effects of mutation may be diminished or abolished by a suppressor mutation. Suppressor mutation is a mutation at a different site from that of the original mutation. A suppressor mutation masks or compensates for the effects of the initial mutation, but it does not reverse the original mutation.
Related Topics