Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Vaccines
Reverse Vaccinology
Reverse Vaccinology
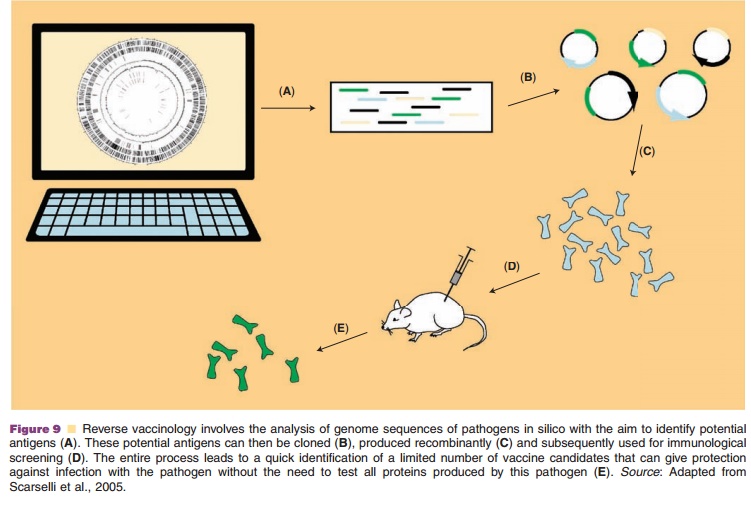
Nowadays vaccines can be designed based on the information encoded by
the genome of a particular pathogen (Masignani et al., 2002; Rappuoli and
Covacci, 2003). From many pathogens the entire genomes have been sequenced and
this number is increasingly growing (http://www.sanger.ac.uk/
Projects/Pathogens). The genome sequence of a pathogen provides a complete
picture of all proteins that can be produced by the pathogens at any given
time. Using computer algorithms, proteins that are either excreted or expressed
on the surface of the pathogen, and thus most likely available for recogni-tion
by the host’s immune system, can be identified. After recombinant production
and purification, these vaccine candidates can be screened for immunogeni-city
in mice. From these, the best candidates can be selected and used as subunit
vaccines (Fig. 9).
A big advantage of reverse vaccinology is the ease at which novel candidate antigens can be selected without the need to cultivate the pathogen. Furthermore, by comparing
genomes of different strains of a pathogen, conserved antigens can be
identified that can serve as a “broad spectrum” vaccine, giving protection
against all strains or serotypes of a given pathogen. One drawback of
thisapproach is that it is limited to the identification of protein-based
antigens.
Reverse vaccinology has been successfully used to identify novel
antigens for a variety of pathogens, including Neisseria meningitidis, Bacillus
anthracis, Streptococcus pneumoniae, Staphylococcus aureus, Chlamydia
pneumoniae and Mycobacterium tuberculosis.
Related Topics