Chapter: Basic Concept of Biotechnology : Biotechnology in Forensic Sciences
Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism was the first technique established to examine variable lengths of DNA fragments formed through DNA digestion. It uses the variations in DNA sequences due to the different locations of restriction enzyme sites. The method practicesthe use of restriction end nucleases to digest the DNA by cutting it at precise sequence arrangements. The resultant restriction fragments are then separated in gel electrophoresis and then shifted to amembrane using the Southern Blot technique. The separated DNA fragments are transferred and probe hybridisation is used to detect the fragments.
In 1980, Wyman and White discovered the first hyper variable locus present in human DNA. The allelic forms identified at this locus fluctuated in size and were so-called restriction fragment length polymorphisms (RFLP's). This procedure became the substance for more exhilarating discoveries.
In 1985, Dr Alec Jeffery’s, using this technology, obtained a DNA fragment that when used as a DNA probe against genomic DNA of a human, attached unambiguously to a number of diverse RFLP sites on the genome generating a distinctive and reproducible banding pattern that was particular for an individual (with the exception of identical twins). He realised this banding pattern was inherited, with half the bands donated from the mother and half from the father. This procedure was named DNA fingerprinting. Originally, DNA fingerprinting was used to define family relationships in immigration applications. However, inSeptember 1986, Dr Jeffery’s was approached by police to use the genetic fingerprinting technology to assist a rape/murder investigation. The highly publicised case, Narborough Murder Enquiry, was the first murder investigation resolved by DNA fingerprinting.
Restriction Enzymes
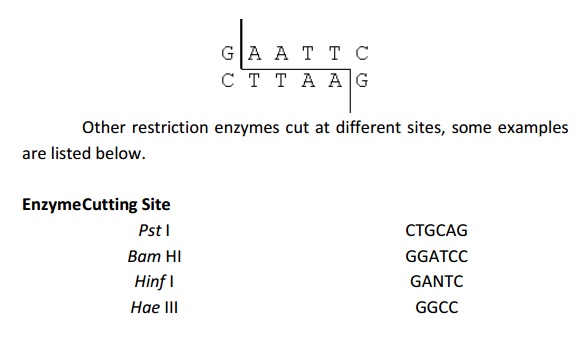
The technique of DNA fingerprinting needs the DNA to be cut up into small fragments. Restriction enzymes perform this digestion. Restriction enzymes were discovered in bacteria, which practice them as a defence mechanism to cut up the DNA of other bacteria or viruses. Hundreds of different restriction enzymes have been extracted. Each one of them cuts DNA at a specific base sequence viz. EcoRI always cuts DNA at GAATTC as indicated below.Other restriction enzymes cut at different sites, some examples are listed below.
In RFLP analysis, restriction enzymes cut up the DNA of an organism into fragments. A large number of short fragments of DNA are produced.
Restriction enzymes cut at the same base sequence always. Since, no two individuals have identical or duplicate DNA; no two individuals would have the same length fragments. For example, the enzyme EcoRI always cuts DNA at the sequence GAATTC (see above). Different people have different numbers of this particular sequence and will consequently have different fragment lengths. Additionally, some of the restriction sites will be at different locations on the chromosome.
The experimental steps being used in forensics laboratory for DNA profile analysis are as follows:
DNA extraction
It is possible to extract DNA from almost any human tissue. Sources of DNA found at a crime sight might include tissue from a deceased victim, blood, cells in a hair follicle, semen and even saliva. The
extracted DNA from the items of evidence is compared to DNA extracted from reference samples i.e. known individuals, normally from blood.
Digestion of DNA with a restriction endonuclease
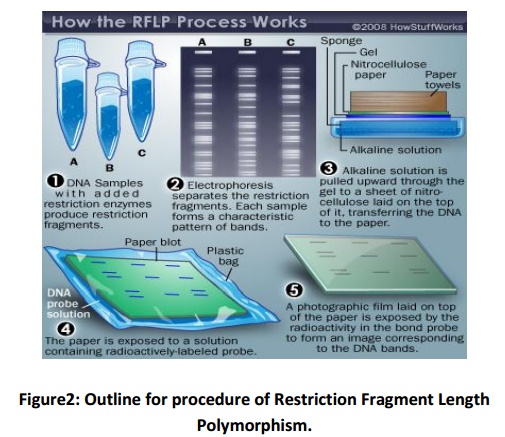
Extracted DNA is exposed to a restriction endonuclease (Fig. 2), which will cut double stranded DNA whenever a specific DNA sequence occurs. The enzyme HaeIIIis most commonly used for forensic DNA analysis, which cuts DNA at the sequence 5'-GGCC-3'.
Agarose gel electrophoresis
The resulting DNA fragments are separated based on size factor via electrophoresis in agarose gels followed by DNA digestion.The migration rate of DNA fragments is slowed by the matrix of the agarose gel. The smaller DNA fragments move faster through the pores of the gel matrix than larger DNA fragments. As a result, a continuous separation of the DNA fragments according to size is obtained with the smallest DNA fragments travelling the greatest distance away from the origin.
Preparation of a Southern blot
After the completion of electrophoresis, the separated DNAs are denatured while still in the agarose gel by soaking the gel in a basic solution. Following neutralization of the basic solution, the single strand DNA molecules are transferred to the surface of a nylon membrane by blotting. After the inventor, Edwin Southern, this denaturation/blotting procedure is known as a Southern blot.The blotting of DNA to a nylon membrane conserves the spatial arrangement of the DNA fragments that occurred after electrophoresis.
Hybridization with radioactive probe
A single locus probe is a DNA or RNA sequence that is able to hybridize (i.e. form a DNA-DNA or DNA-RNA duplex) with DNA from a specific restriction fragment on the Southern blot. The duplex formation depends on the complementary base pairing between the DNA on the Southern blot and the probe sequence. The single locus probes areusually tagged with a radioactive label for an easy detection, and are chosen to detect one polymorphic genetic locus on a single chromosome. Under conditions of temperature and salt concentration that favour hybridization, the Southern blot from step 4 is incubated in a solution containing a radioactive, single locus probe. The unbound probe is washed away after hybridization; consequently, the only radioactivity remaining bound to the nylon membrane is associated with the DNA of the targeted locus.
Detection of RFLPs via autoradiography
Autoradiography detects the locations of radioactive probe hybridization on the Southern blot. In this technique, the washed nylon membrane is overlaid by a sheet of X-ray film in a light tight container. The X-ray film records the locations of radioactive decay. After exposure and development of the X-ray film, the resulting record of the Southern hybridization is termed an autoradiograph, or authored.
Re-probe southern blot with additional probes
After an authored has been developed for the first single locus probe, the radioactivity on the Southern blot can be washed away with a high temperature solution, leaving the DNA in place. The Southern blot can be hybridized with a second radioactive single locus probe, and by repetition of steps 5-7, a series of different single locus probes. The set of autorads from a Southern blot is known as a DNA profile.
Limitations:
However, DNA analysis with RFLP required comparatively large amounts of DNA and degraded samples could not be examined with precision. More effective, quicker and economical DNA profiling techniques have been developed, so RFLP is mostly no longer used in forensic science.
Related Topics