Chapter: Essential Microbiology: Microorganisms in Genetic Engineering
Microorganisms in Genetic Engineering
Microorganisms in Genetic Engineering
Introduction
In the last 30 years or so, there has been a revolution in the
field of genetics, which has had a profound effect on virtually every other
area of biology. This has been due to the development of new techniques that
have enabled scientists to analyse and manipulate DNA in a quite unprecedented
way. Genetically modified crops, DNA ‘fingerprinting’ and gene therapy are just
three of the many applications made possible by these ad-vances.
The beginnings of genetic engineering can be said to date
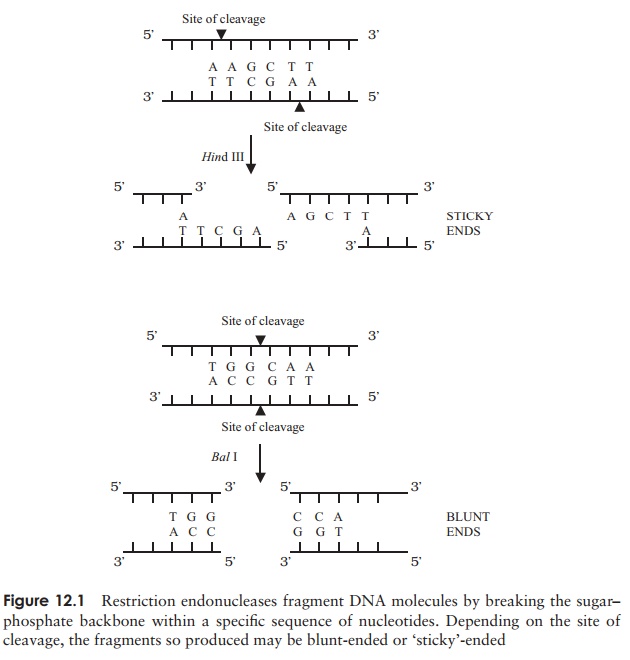
from the discovery, in the late 1960s, of a class of bacterial enzymes called restriction endonucleases (REs). These
are enzymes that cleave DNA into pieces by mak-ing breaks in the
sugar-phosphate backbone; in nature, they serve to destroy any foreign DNA that
may enter the cell. They do not cut the DNA in a random fash-ion, however;
their unique usefulness to the molecu-lar biologist lies in the fact that they
break the DNA in a precise and
reproducible manner. They do this bycutting only at specific recognition sites, sequences of
typically four to six nucleotides (Figure 12.1). Thus, under favourable
conditions, a particular RE will di-gest a given piece of DNA into an identical
collection of fragments, time after time. In the ensuing years, many hundreds
of restriction endonucleases have been discov-ered, many of which recognise
different specific sequences, providing biologists with a hugely versatile tool
for the manipulation of DNA, often likened to a pair of molecular ‘scissors’.
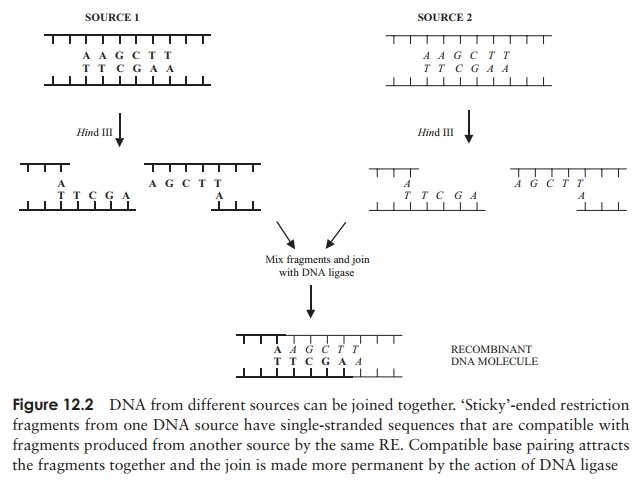
Not long after REs were first isolated, they were used to create the first
man-made recombinant DNA molecule
(Figure 12.2). This involved cutting
fragments of DNA from different sources, then using
another enzyme, DNA ligase to join
them together, a process facilitated by using fragments with compatible ‘sticky’ ends. Remember that
A always pairswith T and C with G; because of this, complementary sequences
that come into contact with one another will ‘stick’ together. DNA, it seems,
is DNA, wherever it comes from; consequently DNA from plants, animals, bacteria
or viruses can be joined together to create novel sequences undreamed of by
Mother Nature.
Of course, a single molecule of our newly recombinant
DNA is not much use to us. The important breakthrough came with the development
of cloning – the ability to produce
huge numbers of copies of a given molecule. Todo this, two further things are
needed: a carrier DNA molecule called a vector,
and a host cell in which it can be
replicated.
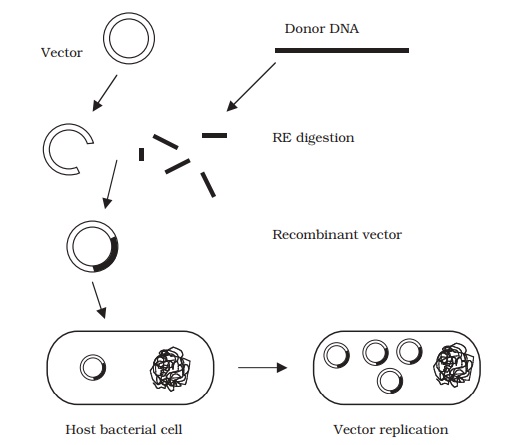
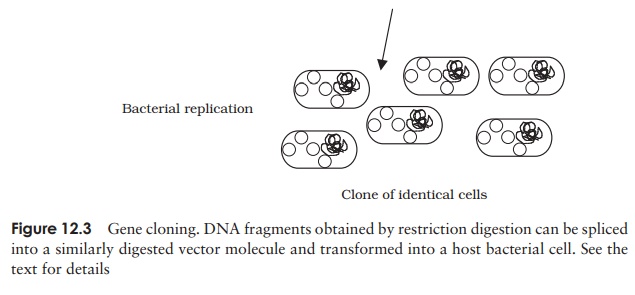
Figure 12.3 shows the main steps of a cloning
protocol:
·
‘donor’ DNA and vector are digested with an RE to
provide compatible sticky ends
·
a fragment of donor DNA is spliced into the vector
molecule
·
the recombinant vector gains entry to a host cell (e.g. E. coli )
·
the vector replicates inside the cell, making further
copies of the inserted DNA
·
host multiplication results in the formation of a
clone of cells, all containing the same recombinant plasmid – we now have
millions of copies of our donor DNA ‘insert’. A collection of such clones is
called a DNA library.
Let us look at role of vectors in a little more
detail. The main features required of a cloning vector are:
it must be
capable of replicating autonomously inside a host cell – when it does
so, any DNA it carries willalso be replicated. Vectors make multiple copies of
themselves inside the host cell.
it must be
relatively small – to facilitate manipulationand entry into a host
cell, vectors must not exceed a certain size.
it must carry a
selectable marker – since only a pro-portion of host cells will take up
the vector, there must be a means of differentiating them from those that do
not. A common way to do this is to use a vector that carries a gene that
confers resistance to an antibiotic such as ampicillin. When bacterial cells
are plated out on a medium containing the antibiotic, only those that have
taken up the vector will be able to form colonies. (The host strain must, of
course, normally be suscep-tible to the antibiotic.)
it must carry a
single copy of RE restriction sites – in order to accommodate a
pieceof donor DNA, a vector must be cut by a restriction endonuclease in one
place only (Figure 12.3).
Related Topics