Chapter: Essential Microbiology: Microorganisms in Genetic Engineering
Bacteriophages as cloning vectors
Bacteriophages
as cloning vectors
The most commonly used bacteriophage vectors are
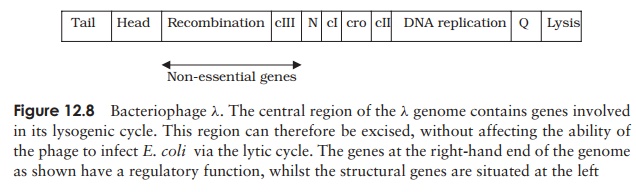
those based on phage Lambda (λ) (Figure 12.8). The genome of λ is 48.5kb in length; it was the first genome to have
its entire sequence determined (1982). As drawn conventionally, the genome is
linear, and contains 46 genes. At each 5 end of the double-stranded structure
is a 12 base single-stranded sequence known as a cos site; these have a complementary base sequence (i.e. they are
cohesive, or ‘sticky’ ends). They can join together to form a double-stranded,
circular λ molecule, a
conformation that is essential for insertion and integration into the host
genome.
The naturally occurring phage is unsuitable as a
vector, because being relatively large, it contains multiple copies of
recognition sites for a number of REs. By using genetic manipulation
techniques, however, unwanted sites can be removed, allowing the DNA to be
‘opened up’ at a single location, and fragments of foreign DNA with
complementary sticky ends to be ligated.
A further limitation that must be overcome is that in
this form, phage λ can only
accommodate about another 3kb of DNA. This is because if the genome exceeds
52kb, it cannot be packaged properly into the protein head to produce viable
phage particles. The arrangement of genes on the phage λ genome offers a solution to this problem. It is
known that genes encoding specific functions are clustered together, with genes
necessary for the lytic replication cycle being found at the ends of the map as
it is conventionally drawn (Figure 12.8). It is possible, therefore, to remove
much of the central part of the λ genome without
affecting its ability to infect and lyse its bacterial host.
Insertion
vectors have had some of this non-essential material removed,
reducing theirgenome size to around 42kb, and thus allowing them to carry an
insert of up to 10kb. A single-copy restriction site is opened up, and a
fragment with complementary sticky ends inserted with the aid of DNA ligase.
Vectors such as λZAPII have a
multiple cloning site or polylinker, positioned so that it falls within the lacZ gene. This allows insertional
Replacement
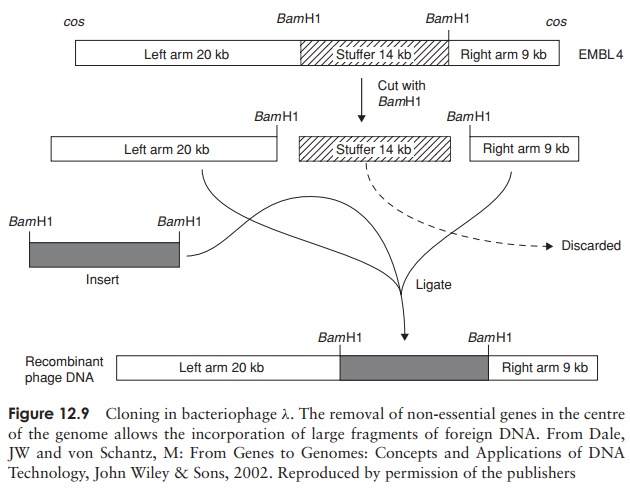
vectors (Figure 12.9) are able to accommodate larger inserts,
becauseinstead of a single copy of a particular RE site, they have two, one
situated either side of a disposable central section known as the ‘stuffer’
fragment. This can be removed by digestion with the appropriate RE and replaced
by insert DNA. The vector λEMBL3, which
can accommodate inserts of between 9 and 23kb, has two polylinker sequences
containing sites for SalI, BamHI and EcoRI flanking the stuffer fragment, allowing a variety of
restriction fragments to be inserted.
Recombinants in replacement vector systems can be
detected by a method that ex-ploits the limitations in the phage’s ability to
package DNA. Just as too big a genome cannot be packaged , neither can one that
is too small (<37kb);
consequently, constructs lacking an insert will not result in the formation of
plaques.
Another naturally occurring bacteriophage of E. coli that has been adapted for use as
a cloning vector is phage M13. This is a single-stranded phage that for part of
its replication cycle inside the host cell exists as a double-stranded
replicative form, which can be isolated and manipulated like a plasmid. Systems
based on M13 have proved popular as a means of obtaining DNA in a
single-stranded form for applications such as DNA sequencing using the dideoxy
(Sanger) method. M13 vectors contain a multiple cloning site situated within
the lacZ gene, allowing blue/white
selection of recombinants. The cloning capacity is generally limited to
fragments of less than 1.5kb, but hybrid M13/plasmid vectors (phagemids) have been developed that are
able to take inserts of as much as 10kb.
As we have seen, a large collection of cloned DNA
fragments is called a DNA library. If the original donor DNA comprised the
entire genome of an organism, then the collection of clones, which between them
contain an entire donor genome, is known as a genomiclibrary. We could create a library of the entire E. coli genome (total size 4.8×106bp)for example,
by having just over 700 clones with an average insert size of 20kb. For a more
complex genome, however, we would either need to have very many more clones, or
to increase the average fragment size.
For this reason, further types of cloning vectors
have been developed, to allow the cloning of larger fragments. Cosmids are entirely man-made creations,
and have no naturally occurring counterpart. They incorporate beneficial
features of both plasmids and phage vectors, and may be capable of
accommodating insert fragments of more than 45kb. Cosmids are essentially
plasmids that contain the cos sites
from λ phage, with
the plasmid supplying the necessary origin of replication, restriction sites
and selectable marker. As we’ve seen, λ DNA will be
packaged into phage heads if the cos
sites are 37–52kb apart; however the only parts of the DNA recognised by the
packaging enzymes are the cos sites,
so any DNA can be used to fill the intervening sequence.
The recombinant DNA is packaged into phage particles
by a process called in vitro
packaging, and introduced into the host E.
coli. Lacking phage genes, cosmids do not lead to lysis of the host cells
and plaque production, but are instead replicated as if they were plasmids.
Growth of host cells on a selective medium allows the detection of
transformants, i.e. those that have taken up the cosmid. There is no need to
select for recombinants, because non-recombinants are too small to be packaged
into the phage heads.
Related Topics