Chapter: Biotechnology: Genomics and Bioinformatics
Microarray Technology
Microarray Technology
It is widely believed that thousands of genes and their products (i.e., RNA and proteins) in a given organism function in a complicated and orchestrated way that creates the mystery of life. However, traditional methods in molecular biology generally work on a "one gene - one experiment" basis, which means that the throughput is very limited and the "whole picture" of gene function is hard to obtain. In the recent years, a new technology, called DNA microarray, has attracted tremendous interests among biologists. This technology promises to monitor the whole genome on a single chip so that researchers can have a better picture of the interactions among thousands of genes simultaneously.
Principle: Microarrays consist of large numbers of DNA molecules spotted in a systematic order on a solid substrate, usually a slide(Fig. 4). The base pairing or hybridization is the underlying principle of DNA microarray. Microarray exploit the preferential binding of complementary single-stranded nucleic acids. A microarray is typically a glass (or some other material) slide, onto which DNA molecules are attached at fixed locations (spots). The type of molecule placed on the array units also varies according to circumstances. The most commonly used molecule is cDNA, or complementary DNA, which is derived from messenger RNA. Since cDNA are derived from a distinct messenger RNA, each feature represents an expressed gene. In order to detect cDNA bound to the microarray, they must be labeled with a reporter molecule that identifies their presence. This technique of introducing fluorescent dyes in DNA and its use in detection of target molecule by hybridization has been previously applied in fluorescent in situ hybridization (FISH).
Procedure: Comparative hybridization experiments compare the amounts of many different mRNA in two cell populations. If one wanted to compare a normal cell and a cancerous cell, the following experiments are needed to be carried out. mRNA is first purified from total cellular contents. mRNA accounts for only about 3% of all RNA in a cell so isolating it in sufficient quantity for an experiment (1-2 micrograms) can be a challenge. Since free RNA is quickly degraded, to prevent the experimental samples from being lost, they are reverse transcribed back into more stable DNA form. The products of this reaction are called complementary (cDNA) because their sequences are the complements of the original mRNA sequences. The reporters currently used in comparative hybridization to microarrays are fluorescent dyes (fluors), represented by the red and green circles attached to the cDNAs in Fig. 4. A differently-colored fluor is used for each sample so that we can tell the two samples apart on the array.
The labelled cDNA samples are called probes because they are used to probe the collection of spots on the array. The two cDNA probes are tested by hybridizing them to a DNA microarray. The array holds hundreds or thousands of spots, each of which contains a different DNA sequence. If a probe contains a cDNA whose sequence is complementary to the DNA on a given spot, that cDNA will hybridize to the spot, where it will be detectable by its fluorescence. In this way, every spot on an array is an independent assay for the presence of a different cDNA. Microarrays are made from a collection of purified DNA molecules typically using an arraying machine. The choice of DNA to be used in the spots on a microarray determines which genes can be detected in a comparative hybridization assay. The end product of a comparative hybridization experiment is a scanned array image. A small piece of such an image is shown in Fig. 4.
Interpretation: The measured intensities from the two fluorescent reporters have been false-coloured red and green and overlaid (in reality, you observe colours when the fluors are stimulated by a laser). Spots, whose mRNA is present at a higher level in one or the other cell population show up as predominantly red or green. Yellow spots have roughly equal amounts of bound cDNA from each cell population. Therefore yellow spots correspond to genes expressed approximately equally in both normal and cancerous cells whereas red spots correspond to genes expressed in high amounts in normal cells. Similarly green spots correspond to genes expressed in high amounts in cancerous cells. By drawing this distinction, we would be able to understand the altered gene expression patterns in cancerous cells. This allows us to further understand the mechanism and make attempts to develop cures.
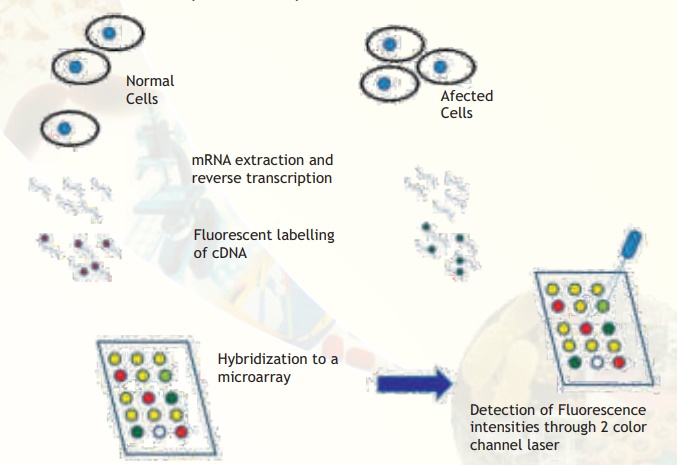
Fig. 4. Major steps involved in comparative microarray hybridization experiments between normal and affected (for example, cancerous) cells are illustrated. Observe that some spots are yellow, meaning that the particular gene is expressed in equal amounts. Some spots are clearly red or green indicating that the particular genes are expressed in only normal or affected condition. Some spots are more greenish or orange meaning that the expression status is not clearly tilted to either side but there is a trend towards either extremities. A few spots may appear blank (no colour).
This microarray technology promises to monitor the whole genome on a single chip so that researchers can have a better picture of the interactions among thousands of genes simultaneously. There are several names to this technology - DNA arrays, gene chips, biochips, DNA chips and gene arrays. In the case of gene chips, the substrate for immobilization is a silicon wafer and the probes are oligonucleotides spotted through photolithographic etching. In this case of gene chip only 1 colour hybridization is performed per chip. Comparisons are done by matching data from one chip to another through a special data normalization procedure. The principle used in this technology is being extended to develop protein arrays also. This technique has been used to study the following:
1. Tissue specific genes
2. Regulatory gene defects in a disease
3. Cellular responses to environment
4. Cell cycle variations
The strength of genomic studies lies in its global comparisons between biological systems. Genomics studies provide initial guidelines to identify areas for deeper investigation and to see how these results fit in the biological context.
Related Topics