Chapter: Biotechnology: Genomics and Bioinformatics
Genome Similarity, SNPs and Comparative Genomics
Genome Similarity, SNPs and Comparative Genomics
The human genome project has often raised two interesting questions - whose genome is being sequenced and how similar are genomes between two different individuals. It is understood that several anonymous samples were collected and pooled for human genome project. Since each person's genome is believed to be 99.8% identical to everyone else's, we can talk in terms of the consensus human genome. SNPs are DNA sequence variations, which occur when a single base (A, C, G, or T) is altered so that different individuals may have different bases at these positions
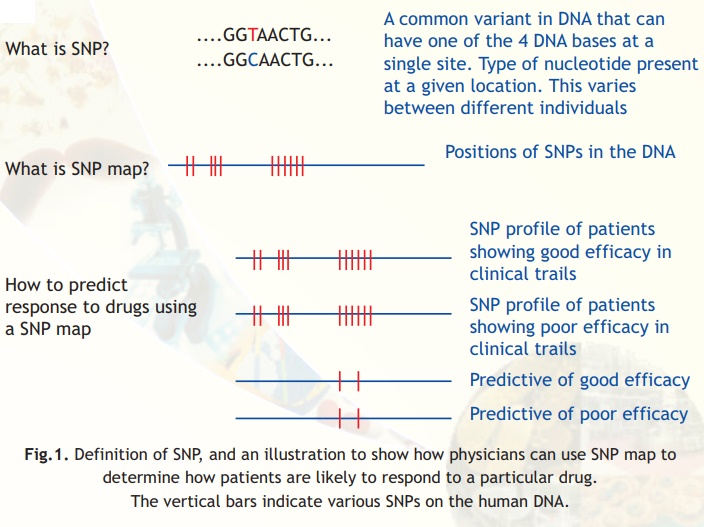
Fig.1. Definition of SNP, and an illustration to show how physicians can use SNP map to determine how patients are likely to respond to a particular drug.
The vertical bars indicate various SNPs on the human DNA.
However, the other way to look at it is that the 0.2% difference in DNA sequence is enough to make each individual unique. It is understood that on an average one in a thousand nucleotides are different in genomes of two different individuals. Particularly, important variations in individual genomes are the single nucleotide polymorphisms or SNPs, which can occur both in coding and non-coding regions of the genome. It is believed that SNPs occur at 1.6 million to 3.2 million sites in the human genome, and may affect gene function, depending upon exact base change and where it occurs. It would be interesting to note the following research based observation.
1. The genetic variations between individuals (particularly, in the non-coding parts of the genome) are exploited in DNA fingerprinting, which is used in forensic science.
2. However, not all genetic variations are beneficial (see Table 2). Genomic variations underlie differences in our susceptibility to, or protection from all kinds of diseases. The severity of illness and the way our bodies respond to treatments are also manifestations of genetic variations. For example, a single base difference in the ApoE gene is associated with Alzheimer's disease, and that a simple deletion within the chemokine-receptor gene CCR5 leads to resistance to HIV (Human Immunodeficiency Virus) infections and the development of AIDS (Acquired Immunodeficiency Syndrome). SNP analysis is therefore important for diagnostics and a SNP database has been developed to aid these applications. An example of how a physician can decide if a medicine prescribed will be effective to a patient is illustrated in Fig. 1 (vertical bars denote various SNPs in patients' genomes).
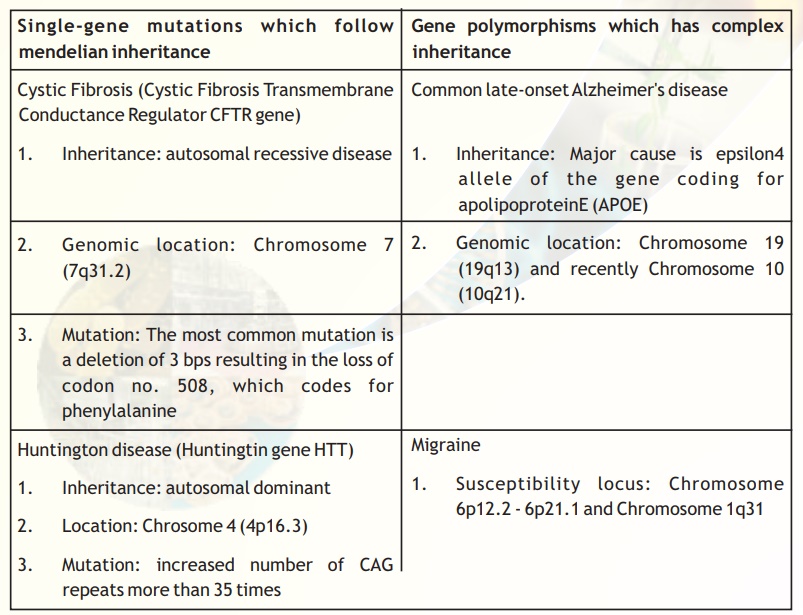
Table 2.Genes and diseases
3. Physicians can use patients DNA sample to determine the pattern of SNP genotype profile and from that they can predict how patients are likely to respond to a particular drug. SNP analysis can also be used in population genetics, as some SNPs vary in different frequencies between populations.
4. The genome sequencing projects have revealed that the genomes of organism otherwise quite different in appearance are quite similar for example mouse and man, are quite similar. Another example is that, among the conserved elements between different species such as the worm and the yeast, substantial portion belongs to genomic regions coding for proteins
5. It is estimated that the difference between human and chimpanzee genomes is only 1 to 3%, while human and mouse share about 97.5% of their working DNA. These similarities suggest that none of these genomes has changed much since we shared a common ancestor 100 million years ago.
Related Topics