Chapter: Biotechnology: Genomics and Bioinformatics
Fluorescence in situ hybridization
Fluorescence in situ hybridization
It is possible to introduce colours into DNA by a technique called Nick Translation developed in 1977 by Rigby and Paul Berg. The enzymes, DNA polymerase I makes DNA and DNase I, which cuts DNA are combined in a buffered reaction with dNTP's, including dUTP labelled with a red or green fluorescence. The DNA polymerase I adds nucleotide residues to the 3-prime hydroxyl terminus that is the result of nicks (breaks) created by the DNase I in the DNA. In the process, the fluorescence labelled nucleotide in the free nucleotide mixture becomes incorporated into the newly synthesized strands of DNA (Fig. 2) .
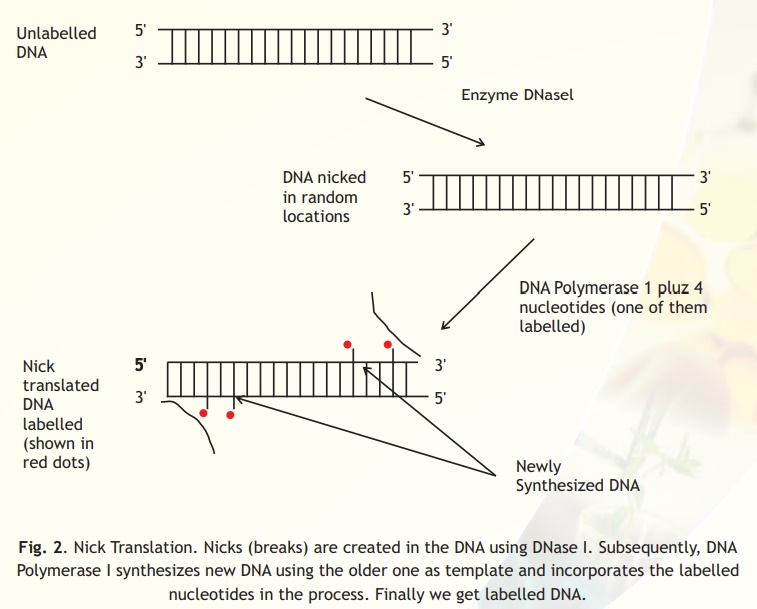
Fig. 2. Nick Translation. Nicks (breaks) are created in the DNA using DNase I. Subsequently, DNA Polymerase I synthesizes new DNA using the older one as template and incorporates the labelled nucleotides in the process. Finally we get labelled DNA.
The DNA fragment size with fluorescence probe after nick translation depends upon thamount of enzyme and the incubation time of reaction. The size range can be 300 to 3000 bp. The application of FISH can be illustrated by taking an example of chronic mylogenous leukemia (CML). It was observed from the karyotype analysis of the lymphocyte preparation made from blood samples of CML patients that there was a 9-22 translocation in the chromosome (also called 'Philadelphia chromosome'). Although by counting the number of such cells it was possible to find out the severity of the disease, it was not an easy procedure. The regions on the chromosomes involved in translocation were identified on chromosomes 9 and 22. From the DNA library it was possible to pick up clones carrying the particular genes involved in CML. Using nick translation it was possible to flourescently label chromosome 9 region with red colour and chromosome 22 region with green colour and prepare the probe (Fig. 3).
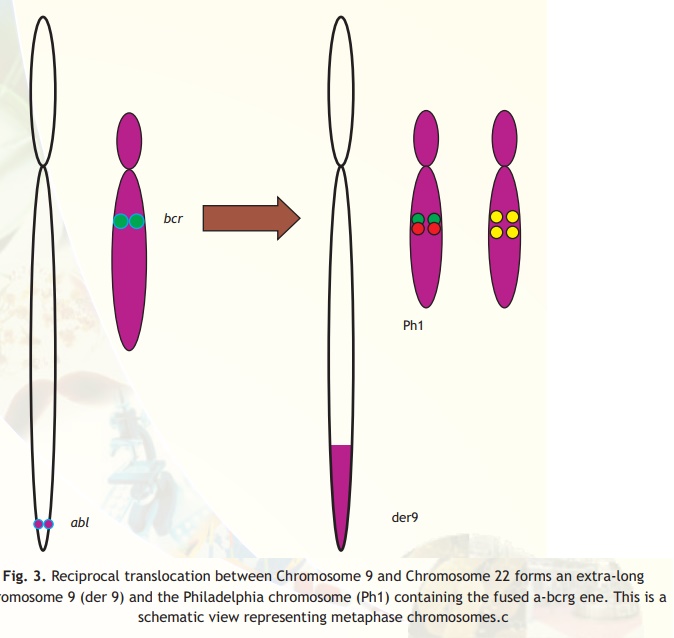
Fig. 3. Reciprocal translocation between Chromosome 9 and Chromosome 22 forms an extra-long chromosome 9 (der 9) and the Philadelphia chromosome (Ph1) containing the fused a-bcrg ene. This is a schematic view representing metaphase chromosomes.c
It was observed that when CML lymphocytes smear cells were hybridized with the two probes in situ and when observed under fluorescent microscope, the cells, which were affected, appeared yellow (mixing of green and red colour produces yellow colour). The unaffected cells appeared as red and green (Fig. 3). This technique known as Fluorescence in situ Hybridization (FISH) allows knowing the status in the interphase unlike in karyotyping where you need a metaphase chromosome. The status of the disease could easily be identified by counting the number of cells, which appeared yellow. Further, it was possible to monitor the effect of chemotherapy and drugs by taking out samples and counting the number of cells appearing
Related Topics