Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Principles of Laboratory Diagnosis of Infectious Diseases
Methods of Nucleic Acid Analysis
Methods of Nucleic Acid Analysis
Agarose Gel Electrophoresis
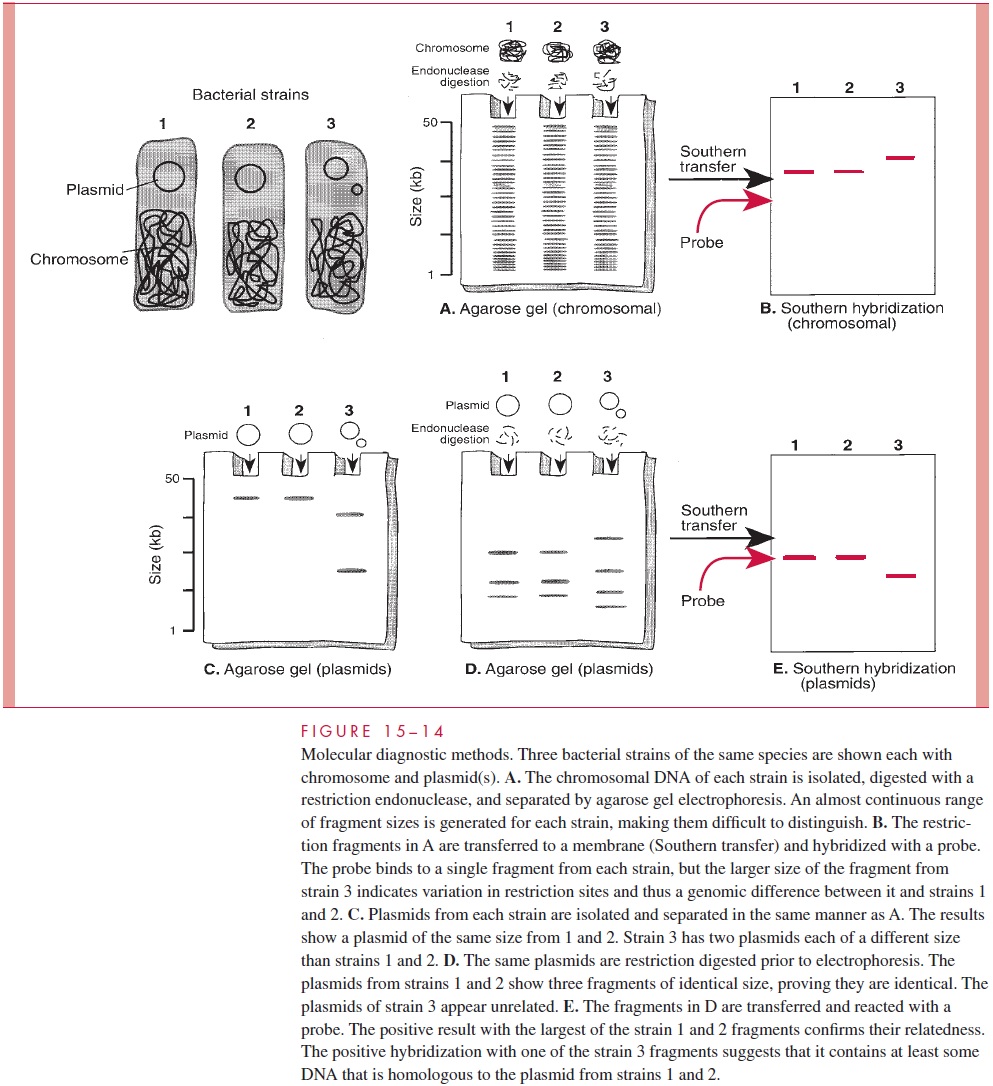
Nucleic acids may be separated in an electrophoretic field in an agarose (highly purified agar) gel. The speed of migration depends on size, with the smaller molecules moving faster and appearing at the bottom (end) of the gel. This method is able to separate DNA fragments in the range of 0.1 to 50 kilobases, which is far below the size of bacterial genomes but in-cludes some naturally occurring genetic elements such as bacterial plasmids (Fig 15 – 14). A variant of agarose gel electrophoresis, pulsed field electrophoresis, alternates the orienta-tion of electrical field in a fashion that allows resolution of much larger DNA fragments.
Restriction Endonuclease Digestion
Restriction endonucleases are enzymes that recognize specific nucleotide sequences in DNA molecules and digest (cut) them at all sites at which the sequence appears. A large number of these enzymes have been isolated from bacterial strains and are commercially available together with information on the sequences they recognize. While the four- to eight-base pair sequences recognized by these endonucleases are not unique to any one organism, their spacing along the chromosome or other genomic structure may be. The size of fragments generated by endonuclease digestion of DNA molecules may be com-pared by agarose gel electrophoresis (Fig 15 – 15).
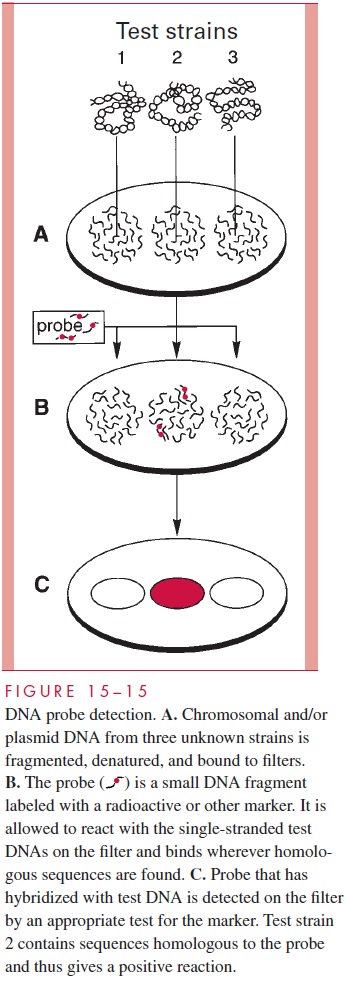
DNA Hybridization
If the DNA double helix is opened, leaving single-stranded (denatured) DNA, the nu-cleotide bases are exposed and thus available to interact with other single-stranded nucleic acid molecules. If complementary sequences of a second DNA molecule are brought into physical contact with the first, they will hybridize to it, forming a new double-stranded mol-ecule in that area. A variety of methods are in use that allow hybridization to take place be-tween two or more nucleic acid molecules. The reaction mixtures vary from tiny probes to the entire genome of an organism. Most immobilize the single-stranded target DNA on a membrane to prevent it from rehybridizing with its own complementary strand, but liquid phase assays have also been developed. A variant in which the DNA is separated by agarose gel electrophoresis before binding to the membrane is called Southern hybridization.
Polymerase Chain Reaction
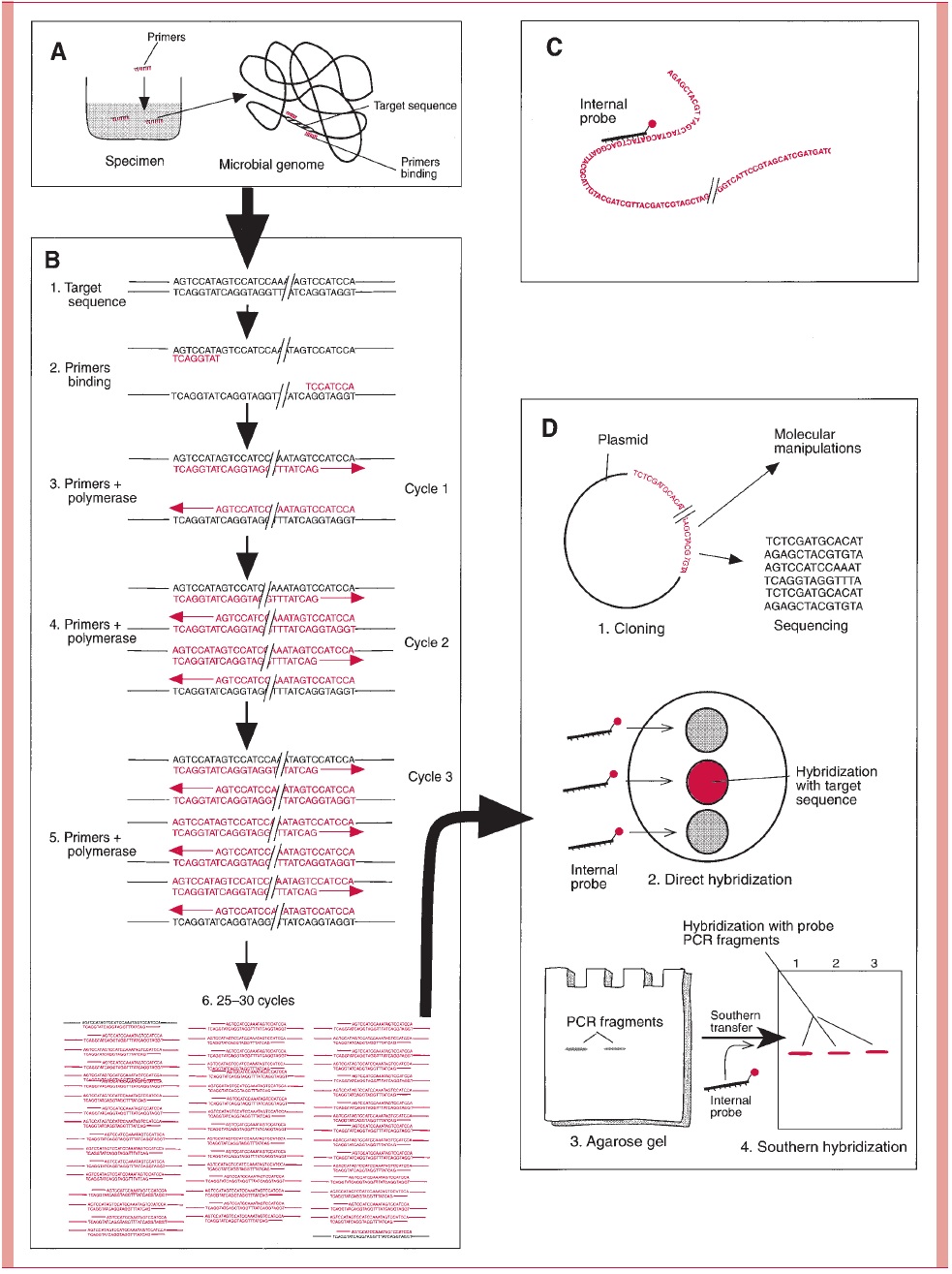
The polymerase chain reaction (PCR) is an amplification technique that allows the detec- tion and selective replication of a targeted portion of the genome. The technique uses special DNA polymerases that through alternate changes in test conditions such as temperature can be manipulated to initiate replication in either the 3or 5direction. The specificity is provided by primers that recognize a pair of unique sites on the chromosome so that the DNA between them can be replicated by repetitive cycling of the test condi- tions. Because each newly synthesized fragment can serve as the template for its own replication, the amount of DNA doubles exponentially with each cycle (Fig 15 – 16).
Nucleic Acid Sequence Analysis
For some time, it has been possible to chemically determine the exact nucleotide se-quence of genomic segments or cloned genes. Published sequences are systematically entered into computer databases such as GenBank and are widely available for analysis by computer software designed to solve a wide variety of problems. Conversely, given the known sequence, short segments of DNA can be synthesized for use as probes or primers. It is even possible to compare a sequenced gene or putative probe against all known se-quences using the computer, an “experiment” that would be impossible in the laboratory.
Related Topics