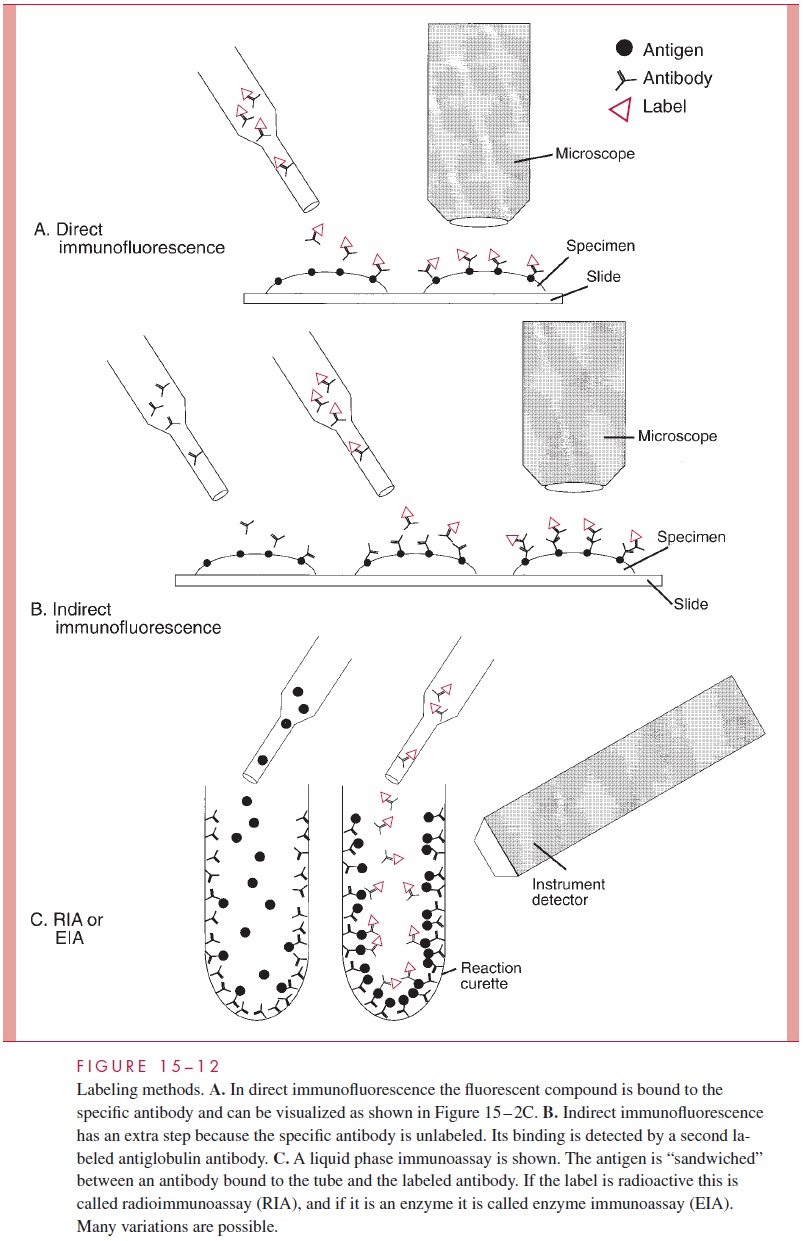
Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Principles of Laboratory Diagnosis of Infectious Diseases
Application of Nucleic Acid Methods to Infectious Diseases
Application of Nucleic Acid Methods to Infectious Diseases
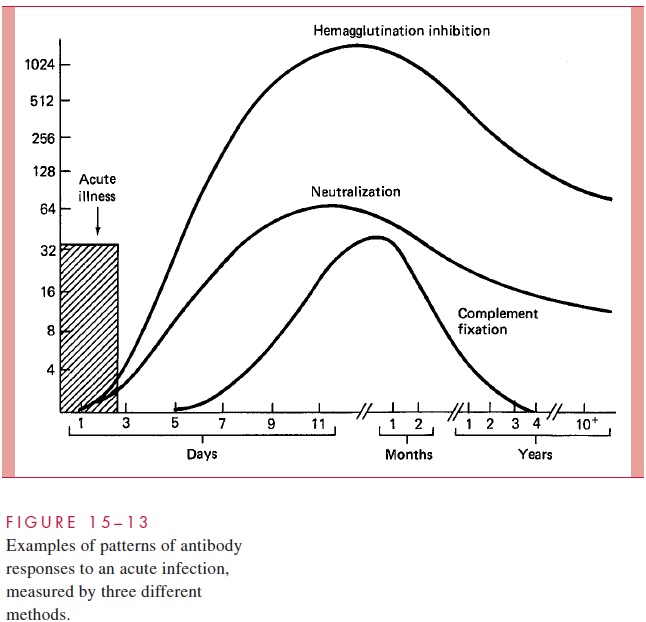
Bacterial and Viral Genomic Sizing
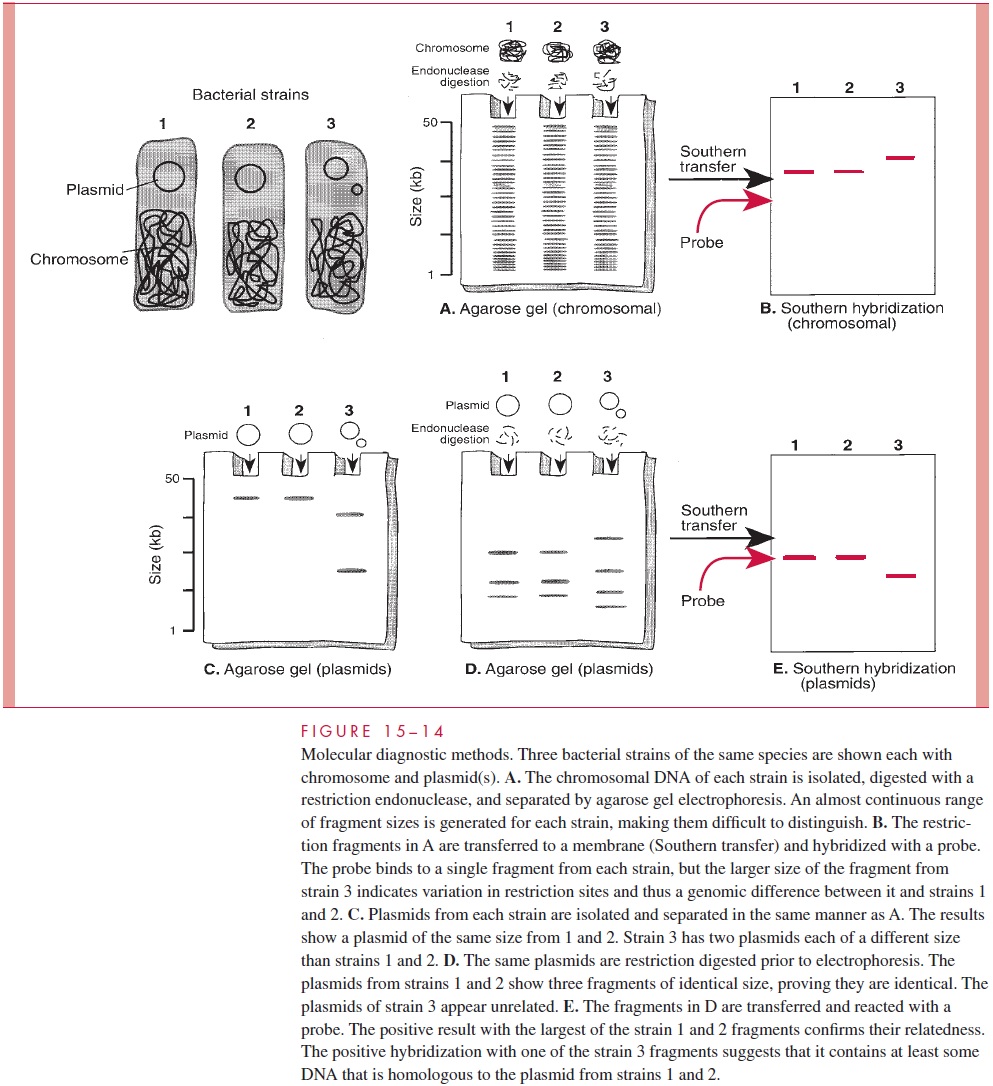
The only intact genetic elements of infectious agents that are small enough to be directly detected and sized by agarose gel electrophoresis are bacterial plasmids. Not all bacterial species typically harbor plasmids, but those that do may carry one or a number of plasmids ranging in size from less than 1 to over 50 kilobases. This diversity makes the presence or absence, number, and sizes of plasmids of considerable value in differentiating strains for epidemiologic purposes. Because plasmids are not stable components of the bacterial genome, plasmid analysis also has the element of a timely “snapshot” of the circumstances of a disease outbreak. The specificity of these results can be improved by digesting the plasmids with restriction endonucleases prior to electrophoresis. Two plasmids of the same size from different strains may not be the same, but if an identical pattern of fragments is generated from the digestion, they almost certainly are. These principles are illustrated Figure 15–14 and their application to an outbreak is shown in Figures 4–12 and 4–13.
Because of their larger size, the chromosomes of bacteria must be digested with endonucleases to resolve them on gels. For viruses the outcome is much like that with plasmids, depending on the genomic size and the endonuclease used. Digested bacterial chromosomes can be compared in this manner, but the number of fragments is very large and the patterns complex. The combined use of endonucleases, which make infrequent cuts, and pulsed-field electrophoresis can produce a comparison comparable to that possible with plasmids. This approach is also used for analysis of the multiple chromosomes of fungi and parasites.
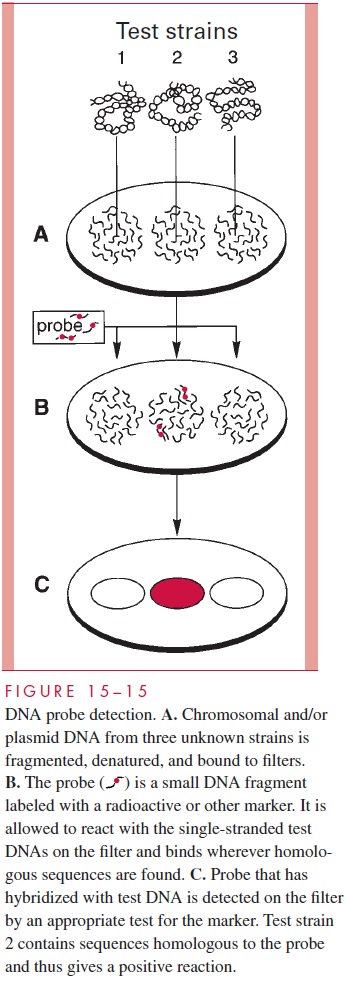
DNA Probes
A “probe” is a fragment of DNA that has been cloned or otherwise recovered from a ge-nomic or plasmid source. It may contain a gene of known function or simply sequences empirically found to be useful for the application in question. In some cases, the probe is synthesized as a single chain of nucleotides (oligonucleotide probe) from known se-quence data. The probes are labeled with a radioisotope or other marker and used in hy-bridization reactions either to detect the homologous sequences in unknown specimens (see Fig 15 – 15) or to further refine gel electrophoresis findings (see Fig 15 – 14). In the latter instance, Southern hybridizations are used to retain knowledge of the size of frag-ments involved. For example, the information that the same gene is present in each of two strains but in different size restriction fragments is evidence for a genomic difference be-tween the two strains (Fig 15 – 14B).
The diagnostic use of DNA probes is to detect or identify microorganisms by hy-bridization of the probe to homologous sequences in DNA extracted from the entire or-ganism. A number of probes have been developed that will quickly and reliably identify organisms already isolated in culture. The application of probes for detection of infec-tious agents directly in clinical specimens such as blood, urine, and sputum is more diffi-cult, because most of the systems developed to date are not as sensitive as culture and are more expensive. However, this approach offers the potential for rapid diagnosis and the detection of characteristics not possible by routine methods. For example, a bacterial toxin gene probe can demonstrate both the presence of the related organism and its toxi-genicity without the need for culture.
Applications of Polymerase Chain Reaction (PCR)
The amplification power of the PCR offers a solution for the sensitivity problems inherent in the direct application of probes (Fig 15 – 16). Although the nucleic acid segment ampli-fied by PCR can be seen directly on a gel, the greatest sensitivity and specificity are achieved when probe hybridization is carried out following PCR. This approach has been successful for a wide range of infectious agents and awaits only further resolution of practical problems for wider use.
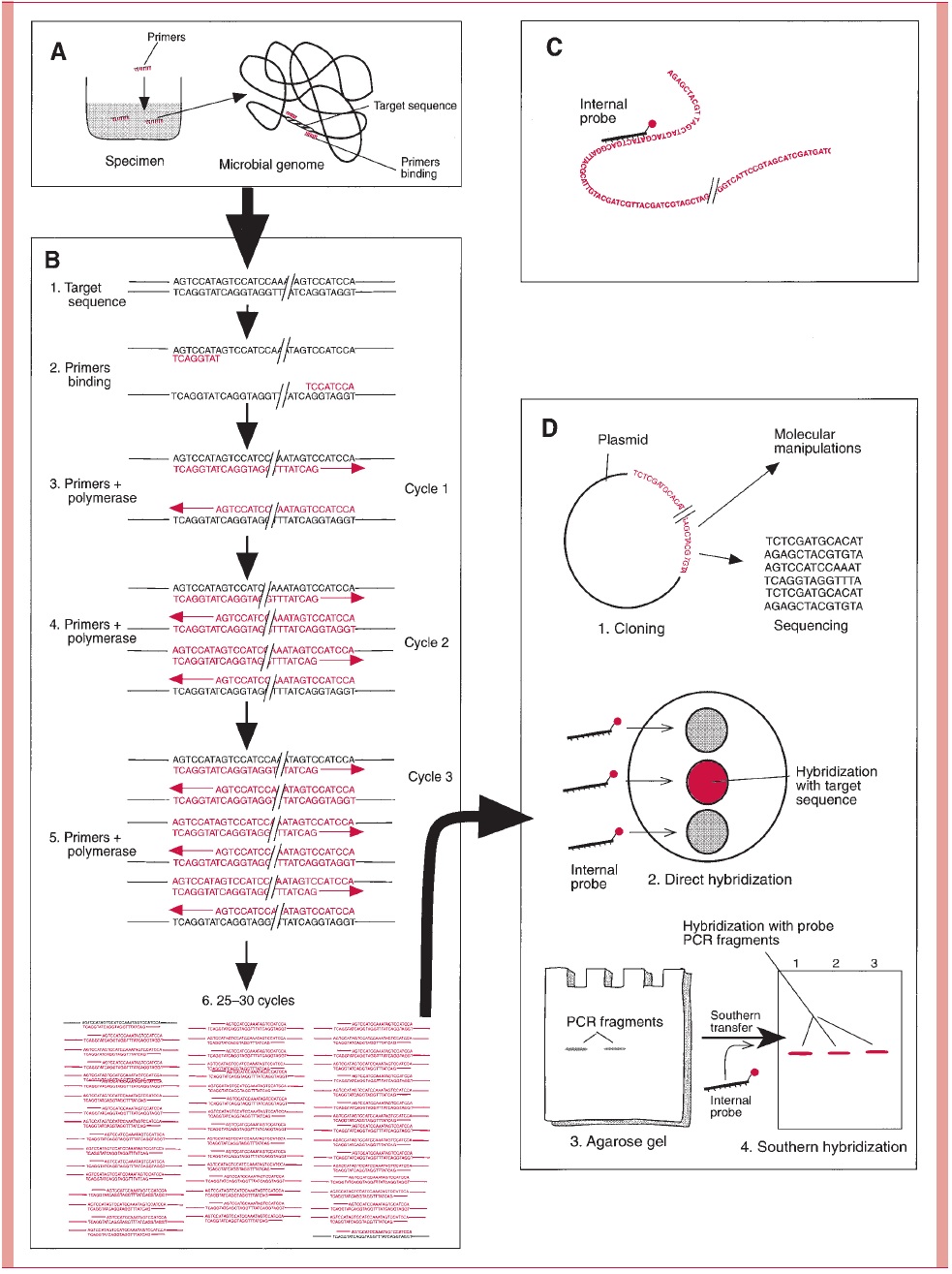
Another creative use of PCR has been in the study of infectious agents seen in tissue but not grown in culture. PCR primers derived from sequences known to be highly con-served among bacteria, such as ribosomal RNA, have been applied to tissue specimens. The amplification produces enough DNA to clone and sequence. This sequence can then be compared with sequences published for other organisms using computers. Thus, taxo-nomic relationships can be inferred for an organism that has never been isolated.
Ribotyping
Ribotyping also makes use of the conserved nature of bacterial ribosomal RNA and of the ability of RNA to hybridize to DNA under certain conditions. Labeled ribosomal RNA of one organism can be hybridized with restriction endonuclease–digested chromosomal DNA of another. In this case, ribosomal RNA is being used as a massive probe of restriction fragments separated by electrophoresis. Hybridization to multiple fragments is common, but if the organisms are genetically different, the restriction fragments, which contain the ribosomal RNA sequences, will vary in size. The pattern of bands produced by epidemiologically related strains can then be compared side by side.
Genomic Analysis
DNA homology techniques hybridize the total genomic DNA of one organism to that of another in a manner demonstrated in Figure 15 – 17. The relatedness of strains can be expressed as a percent homology. Strains related at the species level should show homol-ogy in the 60 to 90% range, whereas strains with increasing taxonomic divergence show progressively less homology.
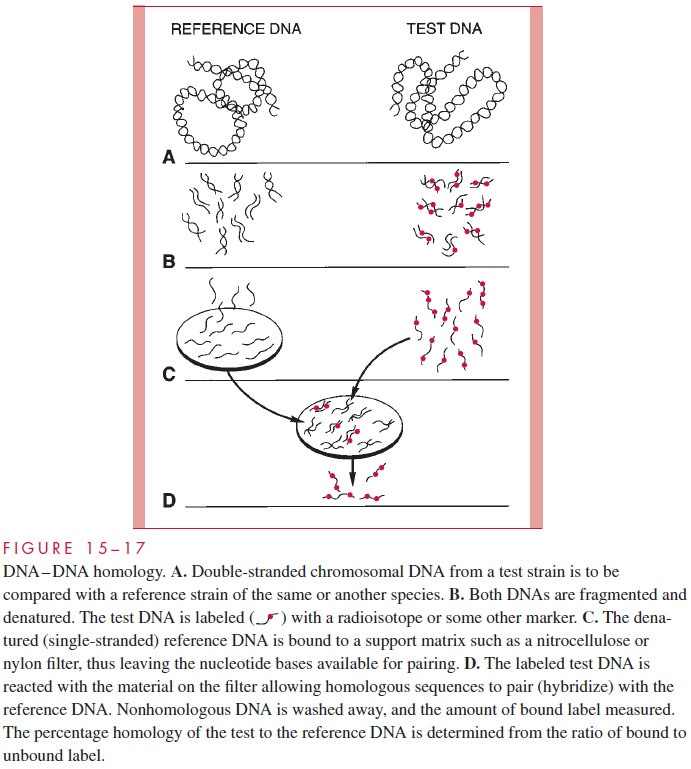
These findings are now a major factor in decisions on the taxonomic classification of all microorganisms, allowing species, genus, and higher taxo-nomic groupings to be assessed by means that are not subject to the phenotypic variation inherent with classical methods. Sequence analysis of mutations in genes related to the action of drugs used in the treatment of AIDS can now be used to study and even predict emergence of drug resistance.
Related Topics