Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Molecular Biotechnology
DNA Sources - Recombinant DNA Technology
DNA Sources
As stated before, any DNA can be used to construct recombinant DNA
molecules. In protein production based on recombinant DNA technology very
distinct pieces of DNA are required. Referring to the metaphor described before
in this chapter, only a few lines on a specific page in one of the many books
are required. Isolation of specific pieces of DNA directly from the DNA of a
bacterial cell and, certainly, of a plant or an animal cell implies a very
tedious search. Can one find a DNA fragment of interest in a more convenient
way? There are several strategies.
Synthetic DNA
It is feasible to use synthetic DNA as a source for the desired
recombinant DNA sequence. If one seeks DNA that codes for a specific protein,
the amino acid sequence of that protein is sometimes known. With the genetic code
as guide one may synthesize the coding DNA by organic synthesis and use that
DNA (called oligonucleotide) to construct an appropriate recombinant DNA
molecule. Although the technique to unravel the amino acid composition of
proteins and the possibilities of the organic DNA synthesis have improved over
the past years, the organic DNA synthesis approach is only feasible to clone
genes coding small proteins. For example, the synthetic approach has been used
successfully for large scale production of human insulin, as will be described
later.
The synthetic DNA approach allows the choice, whenever there are more
triplets available for a certain amino acid, of the triplet that is used most
frequently in the host that is selected for the production. In other words this
technique allows one to master the codon usage problem, mentioned earlier in
this chapter.
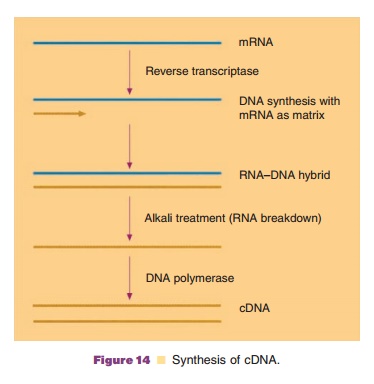
cDNA
An alternative for direct isolation of cellular DNA coding for a
specific protein is the copy DNA (cDNA) approach. This important development in
DNA technology became apparent when the enzyme reverse transcriptase was
exploited as a tool.
Genes are not always expressed in the cell, nor are they expressed
everywhere in the organism. Some genes are only expressed in a certain stage of
cell growth, or under very specific environmental condi-tions, or expressed
only in very specific tissues of an organism. The mRNA molecules in a cell thus
represent a minority of all the available genes, namely those that are actually
expressed. Knowledge of thecell physiology or knowledge of the specific
biological tasks of animal and plant tissues, enables the isolation of a
particular and characteristic set of mRNA molecules. Conversion of these mRNA
molecules by the enzyme reverse transcriptase into DNA leads to synthesis of
the genes that were expressed. These DNA molecules, called cDNA molecules, to
distin-guish them from the natural DNA molecules, can be used for gene cloning.
Provided that the search starts with the right kind of cell culture conditions
or with the appropriate tissues the cDNA strategy can be very efficient. It is
illustrated in Figure 14.
cDNA cloning has some obvious consequences. In contrast to a gene that
is directly isolated from the chromosome, cDNA lacks a promoter region.
Furthermore, in the case of genes harboring exons and introns, cDNA is built
only from exons. The lack of the authentic promoter requires that in the
cloning strategy a promoter should be fused to the cDNA in order to achieve
gene expression. Mostly a strong promoter that can be switched on and off,
depending on environmental or tissue conditions, is fused to cDNA.
Control over foreign gene expression by using a suitable promoter is
very essential as foreign proteins may disturb the physiology of the host cell
and cause a premature stop of cell development. Therefore, a promoter like the lac promoter well-known from molecular pioneer work with Escherichia coli is frequently used in bacterial cells to direct the synthesis of a
foreign protein. This promoter is only switched on when an appropriate inducer
like iso-propyl-b-thiogalactoside (IPTG) is added to the
medium. Cultivation of the cells in the absence of the inducer allows
optimal cell growth since the possible deleterious foreign gene product is not
produced. When high cell numbers are present in the culture, the inducer is
added and consequently the foreign gene product is produced. Possible negative
effects of the foreign protein on the cell are minimal in their consequences
since high cell numbers, often near to the maximum yield, are present when the
harmful production is started.
In an animal or a plant a foreign gene should preferably be expressed
only in certain tissues to keep the animal or the plant vital or to be able to
isolate the protein efficiently. In those biotechnological applica-tions where
mammals like sheep, goat or cow are used as host for pharmaceutical products
(an app-roach called “pharmaceutical farming”), the mam-mary gland is
frequently used as an expression tissue. The cDNA for a product protein is
therefore fused to a promoter that is only expressed in that tissue.
Transferring such a DNA construct into embryos of the host animal may lead to
genetically modified animals that exclusively deliver the pharmaceutical
product as part of their milk. This production route enables an efficient
isolation and allows relatively simple purification strategies.
The lack of introns in the cDNA has the advantage that cDNA may lead to
a functional gene product even in organisms where splicing does not occur (like
in prokaryotic cells) or where splicing is ambiguous or unreliable.
DNA Libraries
The mRNA approach is not feasible if precise knowl-edge of gene
expression is lacking and does not allow cloning of DNA that is not expressed.
A general approach to master very complex DNA molecules for recombinant DNA
technology is to create a DNA library. To do so, random DNA fragments from a
bacterial, plant or animal cell are fused to a vector and then transferred to
an appropriate host. By isolating from a bacterial cell, for example, DNA
fragments that on average amount 1% of total DNA and linking these fragments
individually to a vector, one may create together a few hundred different
recombinant DNA molecules which represent the total bacterial chromo-some.
Using the “DNA-book” metaphor again: the original bacterial “DNA-book” with
about 1000 pages is divided in about 150 small booklets of let say 10 pages
each. These booklets tell, with some overlap and without an a priori ordering, the
same story as told in the complete book. The immediate advantage of this
approach is that large molecules are split in suitable smaller pieces linked to
a replicon.
An individual host with a specific recombinant DNA molecule thus harbors
a fragment of the totalDNA on a replicating vector. By preselecting from the
library the cells carrying gene(s) or DNA of interest, one may obtain a smaller
molecule harboring little more than the DNA of interest by trimming the
fragment isolated from these cells. By subsequent cloning of such smaller
fragments one may achieve the final goal: a recombinant DNA molecule with a
very distinct piece of foreign DNA. It is noteworthy to mention in this respect
that DNA libraries are available from many organisms. Likewise cDNA libraries
exist from many tissues such as the human brain. By analysis of the various
fragments, insight into the genetic structure of all sorts of organisms is
rapidly growing. In this respect the unraveling of the human genome revealed to
the world in the year 2000 is worth mentioning.
Related Topics