Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Molecular Biotechnology
DNA Replication
DNA Replication
Although DNA may be differently organized in various organisms one or more double-stranded DNA molecules in a helix conformation are the predominant structures.
Strands of DNA are com-posed of four specific building elements (shortly
written as A, C, G, and T), the deoxyribonucleotides deoxyadenosine 50-triphosphate, (dATP) deoxy-cytidine 50-triphosphate (dCTP), deoxyguanosine 50-triphosphate (dGTP), and deoxythymidine 50-tri-phosphate (dTTP) linked by phosphodiester bonds. The two strands in
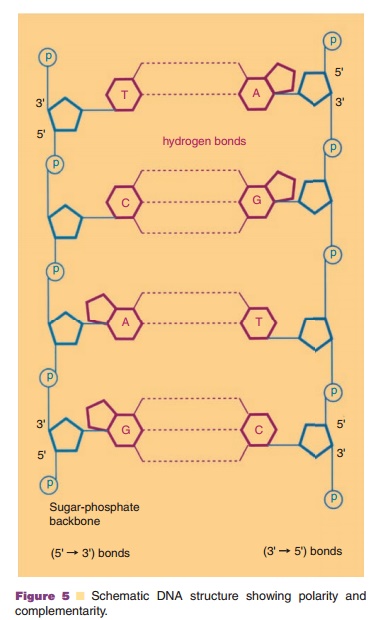
the DNA helix are held together through hydrogen bonds between the nucleotides
in the various strands. The DNA strands in the helix are complementary in their
nucleotide composition: an A in one strand is always facing a T in the other
one, while a C is always facing a G (Fig. 5). Moreover, the strands in
double-stranded DNA run antiparallel: the 50-P end of
the one strand faces the 3 0-OH end
of the complementary strand and the other way round.
During cell division the genetic information in a parental cell is
transferred to the daughter cells by DNA replication. Essential in the very
complex DNA replication process is the action of DNA polymerases. During
replication each DNA strand is copied into a complementary strand that runs
antiparallel. The topological constraint for replication due to the double
helix structure of the DNA is solved by unwinding of the helix, catalyzed by
the enzyme helicase. In a set of biochemical events deoxyribonu-cleotide
monomers are added one by one to the end of a growing DNA strand in a 50 to 30 direction.
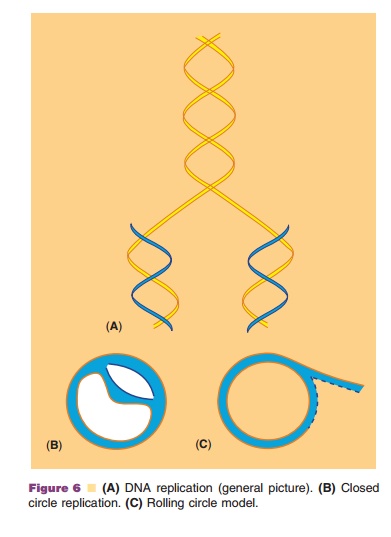
DNA replication starts from specific sites, called origins of
replication (ori). The bacterial chromosome and many plasmids have only one such site.
In the much larger eukaryotic genomes there can be hun-dreds of oris present. For circular DNA molecules like bacterial chromosomes and
plasmids there are two possible ways for the replication. Semi conservative
replication (Fig. 6A) proceeding in the closed circle assuch (Fig. 6B) is one
way. The constraint brought forward by the rotation as a consequence of the
unwinding (there is no free end!) is resolved by the activity of a special
class of enzymes, the topoisome-rases. Alternatively, replication proceeds via
a rolling circle model. In that case the replication starts by cutting one of
the DNA strands in the ori region and then proceeds as
indicated in Figure 6C.
Bacterial plasmids are defined as autonomously replicating DNA
molecules. The basis for that state-ment is the presence of an ori site in the plasmids. The qualification autonomous, however, does not
imply that a plasmid is independent from host factors for replication and
expression. Some plasmids depend on very specific host factors and consequently
they can only replicate in specific hosts. Other plasmids are less specific as
to their host factor requirements and are able to replicate in a broad set of
hosts. As will be demonstrated later, this difference in host range is
meaningful when plasmids are exploited in biotechnology.
Related Topics