Chapter: Biochemistry: Transcription of the Genetic Code: The Biosynthesis of RNA
Transcription in Eukaryotes
Transcription in Eukaryotes
We have
seen that prokaryotes have a single RNA polymerase that is responsible for the
synthesis of all three kinds of prokaryotic RNA-mRNA, tRNA, and rRNA. The
polymerase can switch σ factors to interact with different promoters,
but the core polymerase stays the same. The transcription process is
predictably more complex in eukaryotes than in prokaryotes. Three RNA
polymerases with different activities are known to exist. Each one transcribes
a different set of genes and recognizes a different set of promoters:
·
RNA polymerase I is found in the nucleolus and
synthesizes precursors of most, but not all, ribosomal RNAs.
·
RNA polymerase II is found in the nucleoplasm
and synthesizes mRNA precursors.
·
RNA polymerase III is found in the nucleoplasm
and synthesizes the tRNAs, precursors of 5S ribosomal RNA, and a variety of
other small RNA molecules involved in mRNA processing and protein transport.
All
three of the eukaryotic RNA polymerases are large (500–700 kDa), com-plex
proteins consisting of 10 or more subunits. Their overall structures differ,
but they all have a few subunits in common. They all have two larger subunits
that share sequence homology with the β- and β' -subunits of prokaryotic RNA polymerase that
make up the catalytic unit. There are no σ-subunits to direct polymerases to promoters.
The detection of a gene to be transcribed is accom-plished in a different way
in eukaryotes, and the presence of transcription factors, of which there are
hundreds, plays a larger role. We shall restrict our discussion to
transcription by Pol II.
Structure of RNA Polymerase II
Of the
three RNA polymerases, RNA polymerase II is the most extensively studied, and
the yeast Saccharomyces cerevisiaie
is the most common model system. Yeast RNA polymerase II consists of 12
subunits, as shown in Table 11.2. The subunits are called RPB1 through RPB12. RPB stands for RNA polymerase B because another nomenclature system refers to the
polymerases as A, B, and C, instead of I, II, and III.
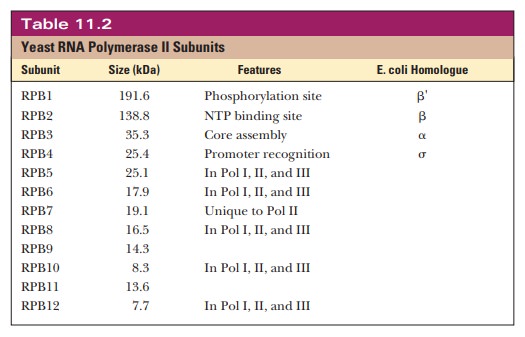
The
function of many of the subunits is not known. The core subunits, RBP1 through
RBP3, seem to play a role similar to their homologues in prokaryotic RNA
polymerase. Five of them are present in all three RNA polymerases. RPB1 has a
repeated sequence of PTSPSYS in the C-terminal
domain (CTD), which, as the name applies, is found at the C-terminal region
of the protein. Threonine, serine, and tyrosine are all substrates for
phosphorylation, which is important in the control of transcription initiation.
X-ray
crystallography has been used to determine the structure of RNA poly-merase II.
Notable features include a pair of jaws formed by subunits RPB1, RPB5, and
RPB9, which appear to grip the DNA downstream of the active site. A clamp near
the active site is formed by RPB1, RPB2, and RPB6, which may be involved in
locking the DNA:RNA hybrid to the polymerase, increasing the stability of the
transcription unit. Figure 11.17 shows a diagram of the structure of RNA
polymerase II.

Recent
structural work on RNA polymerases from prokaryotes and eukary-otes has led to
some exciting conclusions regarding their evolution. Extensive homology exists
between the core regions of RNA polymerases from bacteria, yeast, and humans,
leading researchers to speculate that RNA polymerase evolved
eons ago, at a time when only prokaryotes existed. As more complex organisms
developed, layers of other subunits were added to the core poly-merase to
reflect the more complicated metabolism and compartmentalization of eukaryotes.
How does Pol II recognize the correct DNA to transcribe?
Pol II Promoters
Pol II
promoters have four elements (Figure 11.18). The first includes a variety of upstream elements, which act as enhancers and silencers. Specific binding proteins either activate transcription
above basal levels, in the case of enhancers, or suppress it, in the case of
silencers. Two common elements that are close to the core promoter are the GC
box (–40), which has a consensus
sequence of GGGCGG, and the CAAT box (extending to –110), which has a
consensus sequence of GGCCAATCT.
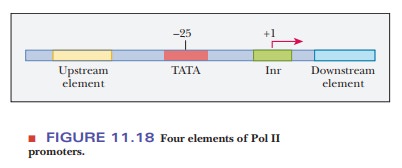
The
second element, found at position –25, is the TATA box, which has a consensus sequence of TATAA(T/A).
The third element includes the transcription start site at position +1, but, in the case of eukaryotes, it is surrounded by a sequence called the initiatorelement (Inr). This sequence is not well conserved. For instance, the sequencefor a particular gene type may be –3YYCAYYYYY+6, in which Y indicates either pyrimidine, and A is the purine at the transcription start site (TSS).
The
fourth element is a possible downstream regulator, although these are more rare
than upstream regulators. Many natural promoters lack at least one of the four
elements. The initiator plus the TATA box make up the core pro-moter and are
the two most consistent parts across different species and genes. Some genes do
not have TATA boxes; they are called “TATA-less” promoters. In some genes, the
TATA box is necessary for transcription, and deletion of the TATA box causes a
loss of transcription. In others, the TATA box orients the RNA polymerase
correctly. Elimination of the TATA box in these genes causes transcription at
random starting points. Whether a particular regulatory ele-ment is considered
part of the promoter or not is often a judgment call. Those that are considered
part of the promoter are close to the TSS (50–200 bp) and show specificity with
regard to distance and orientation of the sequence. Regulatory sequences that
are not considered part of the promoter can be far removed from the TSS, and
their orientation is irrelevant. Experiments have shown that when such
sequences are reversed, they still work, and when they are moved several
thousand base pairs upstream, they still work.
Initiation of Transcription
The
biggest difference between transcription in prokaryotes and eukaryotes is the
sheer number of proteins associated with the eukaryotic version of the process.
Any protein that regulates transcription but is not itself a subunit of RNA
polymerase is a transcription factor.
There are many transcription factors for eukaryotic transcription, as we shall
see. The molecular mass of the entire complex of Pol II and all of the
associated factors exceeds 2.5 million Da.
Transcription
initiation begins by the formation of a preinitiation
complex, and most of the control of transcription occurs at this step. This
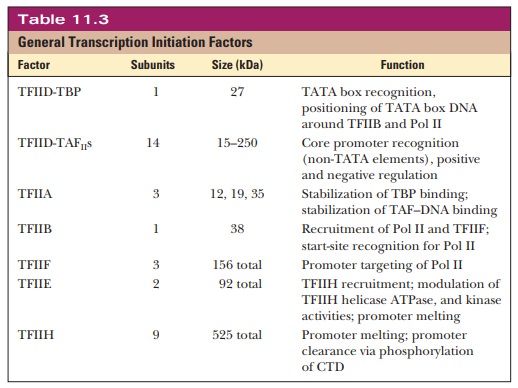
complex normally contains RNA polymerase II and six general transcription factors(GTFs)-TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH.
What do eukaryotic transcription factors do?
The
general transcription factors are required for all promoters. Much work is
still going on to determine the structure and function of each of the parts of
the preinitiation complex. Each GTF has a specific function, and each is added
to the complex in a defined order. Table 11.3 is a summary of the components of
the preinitiation complex.
Figure
11.19 shows the sequence of events in Pol II transcription. The first step in
the formation of the preinitiation complex is the recognition of the TATA box
by TFIID. This transcription factor is actually a combination of several
proteins. The primary protein is called TATA-binding
protein (TBP). Associated with TBP are many TBP-associated factors (TAFIIs). Because TBP is also present and required
for Pol I and Pol III, it is a universal transcription factor. TBP is highly
conserved. From species as different as yeast, plants, fruit flies, and humans,
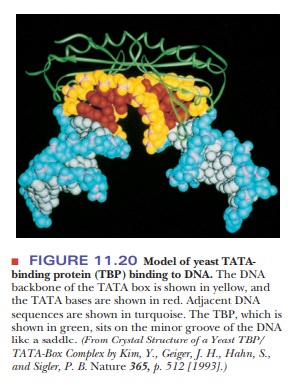
the TBPs have more than 80% identical amino acids. The TBP protein binds to the
minor groove of the DNA at the TATA box via the last 180 amino acids of its
C-terminal domain. As shown in Figure 11.20, the TBP sits on the TATA box like
a saddle. The minor groove of the DNA is opened, and the DNA is bent to an 80°
angle.
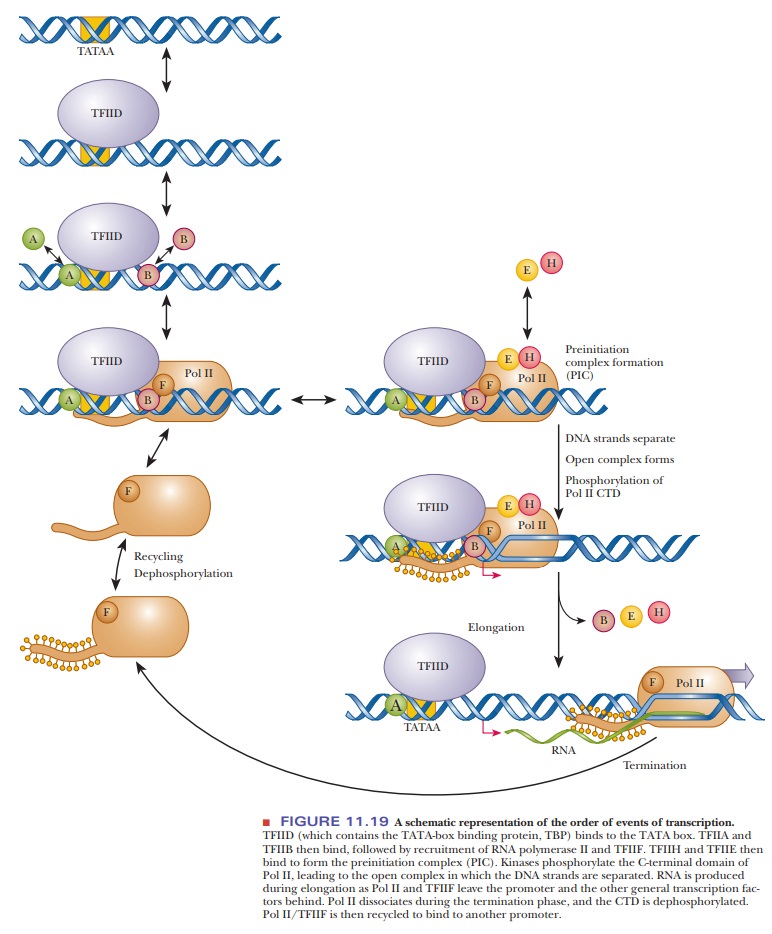
As shown in Figure 11.19, once TFIID is bound, TFIIA binds, and TFIIA also interacts with both the DNA and TFIID. TFIIB also binds to TFIID, bridging the TBP and Pol II. TFIIA and TFIIB can bind in either order, and they do not interact with each other. TFIIB is critical for the assembly of the initiation com-plex and for the location of the correct transcription start site. TFIIF then binds tightly to Pol II and suppresses nonspecific binding. Pol II and TFIIF then bind stably to the promoter. TFIIF interacts with Pol II, TBP, TFIIB, and the TAFIIs. It also regulates the activity of the CTD phosphatase.
The last
two factors to be added are TFIIE and TFIIH. TFIIE interacts with
unphosphorylated Pol II. These two factors have been implicated in the
phos-phorylation of polymerase II. TFIIH also has helicase activity. After all
these GTFs have bound to unphosphorylated Pol II, the preinitiation complex is
complete. TFIIH has been found to have other functions as well, such as DNA
repair.
Before
transcription can begin, the preinitiation complex must form the open complex. In the open complex, the
Pol II CTD is phosphorylated, and theDNA strands are separated (Figure 11.19).
Elongation and Termination
Less is
known about elongation and termination in eukaryotes than in prokaryotes. Most
of the research efforts have focused on the preinitiation complex and on the
regulation by enhancers and silencers. As shown in Figure 11.19, the
phosphorylated Pol II synthesizes RNA and leaves the promoter region behind. At
the same time, the GTFs either are left at the promoter or dissociate from Pol
II.
Pol II
does not elongate efficiently when alone in vitro. Under those circum-stances,
it can synthesize only 100–300 nucleotides per minute, whereas the in vivo
rates are between 1500 and 2000 nucleotides per minute. The difference is due
to elongation factors. One is TFIIF, which, in addition to its role in the
formation of the preinitiation complex, also has a separate stimulatory effect
on elongation. A second elongation factor, which was named TFIIS, was more recently discovered.
Elongation is controlled in several ways. There are sequences called pausesites, where the RNA polymerase hesitates. This is very similar to the transcrip-tion attenuation we saw with prokaryotes. Elongation can also be aborted, leading to premature termination.
Finally, elongation can proceed past the normal
termi-nation point. This is called antitermination.
The TFIIF class of elongation factors promotes a rapid read-through of pause
sites, perhaps locking the Pol II into an elongation-competent form that does
not pause and dissociate.
The
TFIIS class of elongation factors are called arrest release factors. They help the RNA polymerase move again
after it has paused. A third class of elongation factors consists of the P-TEF and N-TEF proteins (positive-transcription
elongationfactor and
negative-transcription elongation factor). They increase the productiveform
of transcription and decrease the abortive form, or vice versa. At some point
during either elongation or termination, TFIIF dissociates from Pol II.
Termination
begins by stopping the RNA polymerase. There is a eukaryotic consensus sequence
for termination, which is AAUAAA. This sequence may be 100–1000 bases away from
the actual end of the mRNA. After termination occurs, the transcript is
released, and the Pol II open form (phosphorylated) is released from the DNA.
The phosphates are removed by phosphatases, and the Pol II/TFIIF complex is
recycled for another round of transcription (Figure 11.19).
Summary
Eukaryotic transcription is far more complicated than the
prokaryotic version.
There are three RNA polymerases in eukaryotes, of which Pol II
produces mRNA.
Pol II is a large protein with at least 12
subunits. Some of the subunits share homology with the subunits of prokaryotic
RNA polymerase as well as with eukaryotic Pol I and Pol III.
The organization of promoters and enhancers is more complicated
with eukaryotes. An important promoter element is the TATA box at –25.
Initiation
of eukaryotic transcription is also much more complicated. In addition to the
polymerase and the promoter, six general transcription factors are involved in
forming the initiation complex.
Related Topics