Chapter: Biochemistry: Transcription of the Genetic Code: The Biosynthesis of RNA
Transcription Regulation in Prokaryotes
Transcription
Regulation in Prokaryotes
Although
many RNAs and proteins are produced in even a simple prokaryotic cell, not all
of them are produced at the same time or in the same quantities. In
prokaryotes, the control of transcription is largely responsible for
controlling the level of protein production. In fact, many equate transcription
control with gene expression.
How is transcription controlled in prokaryotes?
In prokaryotes, transcription
is controlled in four principal ways-alternative factors, enhancers, operons,
and transcription attenuation. They will be discussed in turn.
Alternative σ
Factors
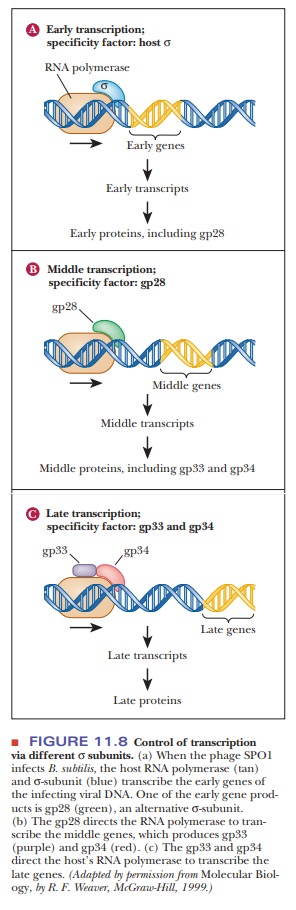
Viruses
and bacteria can exert some control over which genes are expressed by producing
different σ-subunits
that direct the RNA polymerase to different genes. A classic example of how
this works is the action of phage SPO1, a virus that infects the bacteria Bacillus subtilis. The virus has a set
of genes called the early genes,
which are transcribed by the host’s RNA polymerase, using its regular σ-subunit (Figure 11.8). One
of the viral early genes codes for a protein called gp28. This protein is actually another σ-subunit, which directs the RNA polymerase to
transcribe preferentially more of the viral genes during the middle phase. Products of the middle
phase transcription are gp33 and gp34, which together make up another σ factor that directs the
transcription of the late genes. Remember
thatσfactors
are recycled. Initially, the B. subtilis usesthe
standard σ factor.
As more and more of the gp28 is produced, it competes for binding with standard
σ for the
RNA polymerase, eventually subverting the transcription machinery for the virus
instead of the bacterium.
Another
example of alternative σ factors is seen in the response of E. coli to heat shock. The normal σ-subunit in this species is
called σ70 because
it has a molecular weight of 70,000 Da. When E. coli are grown at higher temperatures than their optimum, they
produce another set of proteins in response. Another factor, called σ32, is
produced. It directs the RNA polymerase to bind to dif-ferent promoters that
are not normally recognized by σ70.
Enhancers
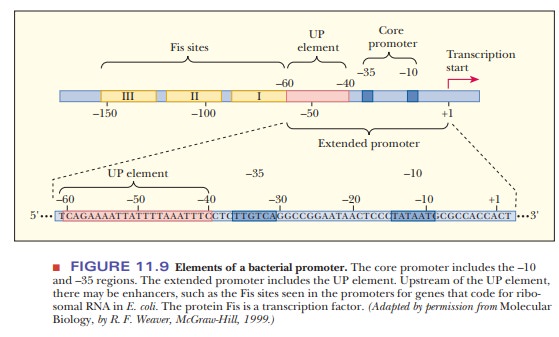
Certain E. coli genes include sequences upstream
of the extended promoter region. The genes for ribosomal RNA production have
three upstream sites, called Fis sites
because they are binding sites for the protein called Fis (Figure 11.9). These
sites extend from the end of the UP element at –60 to –150, and are examples of
a class of DNA sequences called enhancers.
Enhancers are sequences that can be bound by proteins called transcription factors.
What is the difference between an enhancer and a promoter?
When a DNA sequence is labeled a promoter, it implies that the RNA polymerase binds to that region of DNA. An enhancer, on the other hand, is a DNA sequence, usually upstream of the promoter. The polymerase does not bind to enhancers. When enhancers allow a response to changing metabolic conditions, such as temperature shock, they are usually referred to as response elements. When binding the transcription factor increases the level of transcription, the element is said to be an enhancer.
When binding the transcription factor decreases
transcription, the element is said to be a silencer.
The position and orientation of enhancers is less important than for sequences
that are part of the promoter. Molecular biologists can study the nature of
control elements by making changes to them. When enhancer sequences are moved
from one place on the DNA to another or have their sequences reversed, they
still function as enhancers. The study of the number and nature of
transcription factors is the most common research in molecular biology these
days.
Operons
In
prokaryotes, genes that encode enzymes of certain metabolic pathways are often
controlled as a group, with the genes encoding the proteins of the pathway
being close together and under the control of a common promoter. Such a group
of genes is called an operon.
Usually these genes are not transcribed all the time. Rather, the production of
these proteins can be triggered by the presence of a suitable substance called
an inducer. This phenomenon is
called induction. A particularly
well-studied example of an inducible protein is theenzyme β−galactosidase in E. coli.
The
disaccharide lactose (a β-galactoside;) is the
substrate of β-galactosidase.
The enzyme hydrolyzes the glycosidic linkage between galactose and glucose, the
monosaccharides that are the component parts of lactose. E. coli can survive with lactose as its sole carbon source. To do
so, the bacterium needs β-galactosidase to catalyze the first step in
lactose degradation.
The
production of β-galactosidase
takes place only in the presence of lactose, not in the presence of other
carbon sources, such as glucose. A metabolite of lac-tose, allolactose, is the
actual inducer, and β-galactosidase
is an inducible enzyme.
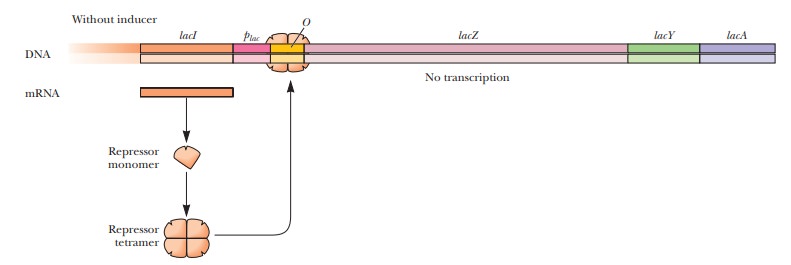
β-Galactosidase is coded for
by astructural gene(lacZ) (Figure 11.10).Structural genes
encode the gene products that are involved in the biochemi-cal pathway of the
operon. Two other structural genes are part of the operon. One is lacY, which encodes the enzyme lactose
permease, which allows lactose to enter the cell. The other is lacA, which encodes an enzyme called
trans-acetylase. The purpose of this last enzyme is not known, but some
hypothesize that its role is to inactivate certain antibiotics that may enter
the cell through
the
lactose permease. The expression of these structural genes is in turn under
control of a regulatory gene (lacI ), and the mode of operation of the
regulatory gene is the most important part of the lac operon mechanism. The regulatory gene is responsible for the
production of a protein, the repressor.
As the name indicates, the repressor inhibits the expression of the structural
genes. In the presence of the inducer, this inhibition is removed. This is an
example of nega-tive regulation because
thelacoperon is turned on unless
something is presentto turn it off, which is the repressor in this case.
How does repression work in the lac operon?
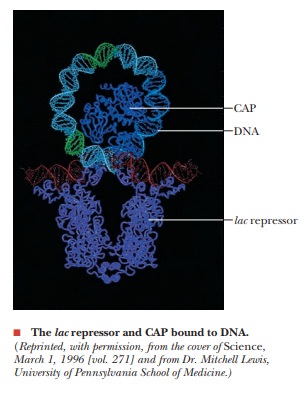
The repressor protein that is made by the lacI gene forms a tetramer when it is translated. It then binds to a portion of the operon called the operator (O) (Figure 11.10). When the repressor is bound to the operator, RNA polymerase cannot bind to the adjacent promoter region (plac), which facilitates the expression of the structural genes. The operator and promoter together constitute the control sites.
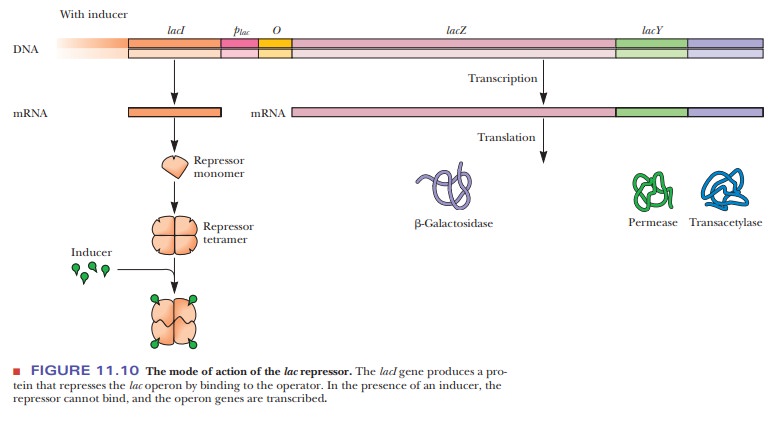
In
induction, the inducer binds to the repressor, producing an inactive repressor
that cannot bind to the operator (Figure 11.10). Because the repres-sor is no longer
bound to the operator, RNA polymerase can now bind to the promoter, and
transcription and translation of the structural genes can take place. The lacI gene is adjacent to the structural
genes in the lac operon, but this
need not be the case. Many operons are known in which the regulatory gene is
far removed from the structural genes.
The lac operon is induced when E. coli has lactose, and no glucose,
avail-able to it as a carbon source. When both glucose and lactose are present,
the cell does not make the lac
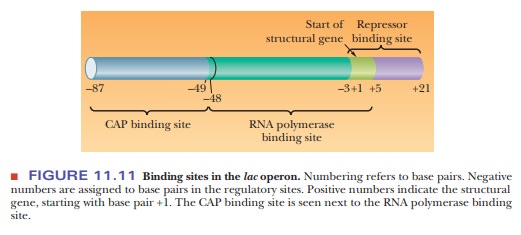
proteins. The repression of the synthesis of the lac proteins by glucose is called catabolite repression. The mechanism by which E. coli recognizes the presence of glucose involves the promoter.
The promoterhas two regions. One is the binding site for RNA polymerase, and
the other is the binding site for another regulatory protein, the catabolite activator protein(CAP) (Figure
11.11). The binding site for RNA polymerase also overlaps thebinding site for
the repressor in the operator region.
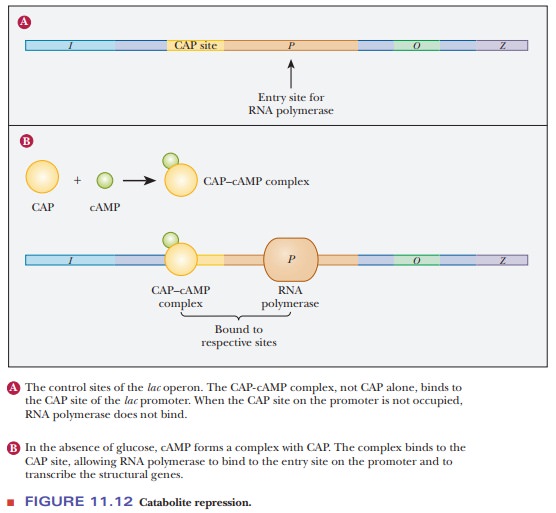
The
binding of CAP to the promoter depends on the presence or absence of
3,'5'-cyclic AMP (cAMP). When glucose is not present, cAMP is formed, serving
as a “hunger signal” for the cell. CAP forms a complex with cAMP. The complex
binds to the CAP site in the promoter region. When the complex is bound to the
CAP site on the promoter, the RNA polymerase can bind at the binding site
available to it and proceed with transcription (Figure 11.12). The lac promoter is particularly weak, and
RNA polymerase binding is minimal inthe absence of the CAP–cAMP complex bound
to the CAP site. The CAP site is an example of an enhancer element, and the
CAP–cAMP complex is a tran-scription factor. The modulation of transcription by
CAP is a type of positiveregulation.
When the cell has an adequate supply of glucose, the level of cAMP is low. CAP binds to the promoter only when it is complexed to cAMP. The combi-nation of positive and negative regulation with the lac operon means that the presence of lactose is necessary, but not sufficient, for transcription of the operon structural genes. It takes the presence of lactose and the absence of glucose for the operon to be active. As we shall see later, many transcription factors and response elements involve the use of cAMP, a common messenger in the cell.
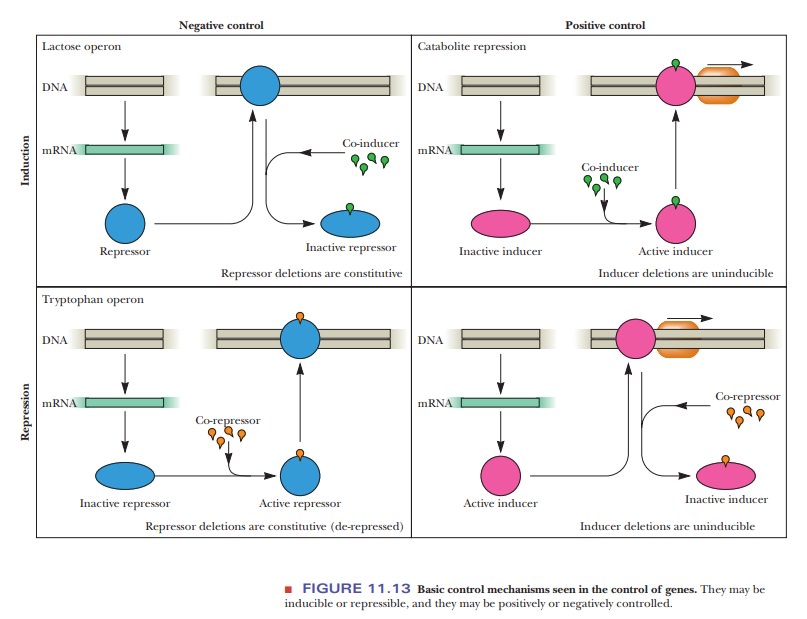
Operons can be controlled by positive or negative regulation mechanisms. They are also classified as inducible, repressible, or both, depending on how they respond to the molecules that control their expression. There are four general possibilities, as shown in Figure 11.13. The top left figure shows a nega-tive control system with induction. It is negative control because a repressor protein stops transcription when it binds to the promoter. It is an inducible system because the presence of the inducer or co-inducer, as it is often called, releases the repression, as we saw with the lac operon. Negative control systems can be identified by the fact that, if the gene for the repressor is mutated in some way that stops the expression of the repressor, the operon is always expressed. Genes that are always expressed are called constitutive. The top right figure shows a positively controlled inducible system. The controlling pro-tein is an inducer that binds to the promoter, stimulating transcription, but it works only when bound to its co-inducer. This is what is seen with the catabolite activator protein with the lac operon. Such positively controlled systems can be identified by the fact that, if the gene for the inducer is mutated, it cannot be expressed-that is, it is uninducible. The bottom left figure shows a negatively controlled repressible system. A repressor stops transcription, but this repres-sor functions only in the presence of a co-repressor. The bottom right figure shows a positively controlled repressible system. An inducer protein binds to the promoter, stimulating transcription; but, in the presence of the co-repressor, the inducer is inactivated.
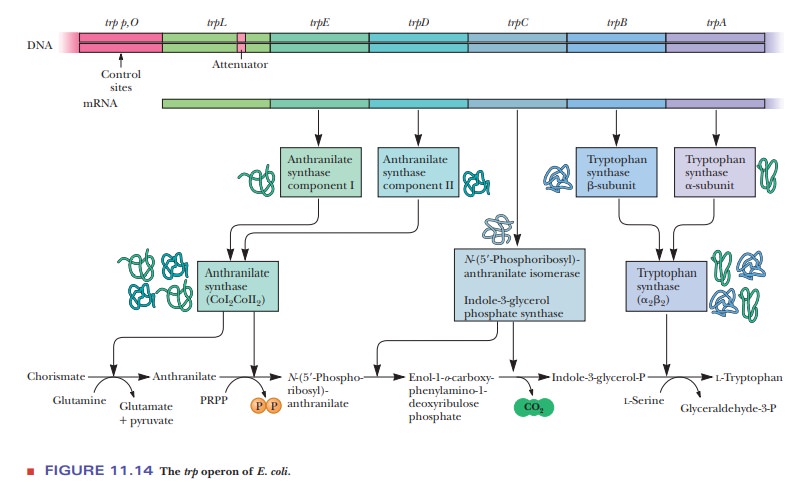
The trp operon of E. coli codes for a leader sequence (trpL) and five polypep-tides, trpE
through trpA, as shown in Figure
11.14. The five proteins make up four different enzymes (shown in the three
boxes near the bottom of the figure). These enzymes catalyze the multistep
process that converts chorisimate to trypto-phan. Control of the operon is via
a repressor protein that binds to two molecules of tryptophan. When tryptophan
is plentiful, this repressor–tryptophan complex binds to the trp operator that is next to the trp promoter. This binding prevents the
binding of RNA polymerase, so the operon is not transcribed. When trypto-phan
levels are reduced, the repression is lifted because the repressor will not
bind to the operator in the absence of the co-repressor, tryptophan. This is an
example of a system that is repressible and under negative regulation, as shown
in Figure 11.13. The trp repressor
protein is itself produced by the trpR
operon and also represses that operon. It is an example of autoregulation, because the product of the trpR operon regulates its own production.
Transcription Attenuation
In addition to repression, the trp operon is regulated by transcriptionattenuation. This control mechanism works by altering transcriptionafterit hasbegun via transcription termination or pausing. Prokaryotes have no separation of transcription and translation as eukaryotes do, so the ribosomes are attached to the mRNA while it is being transcribed.
The trp
operon’s first gene is the trpL sequence
that codes for a leader peptide. This leader peptide has two keytryptophan
residues in it. Translation of the mRNA leader sequence depends on having an
adequate supply of tryptophan-charged tRNA. When tryptophan is scarce, the
operon is translated normally. When it is plentiful, transcription is
terminated prematurely after only 140 nucleotides of the leader sequence have
been transcribed. Secondary structures formed in the mRNA of the leader
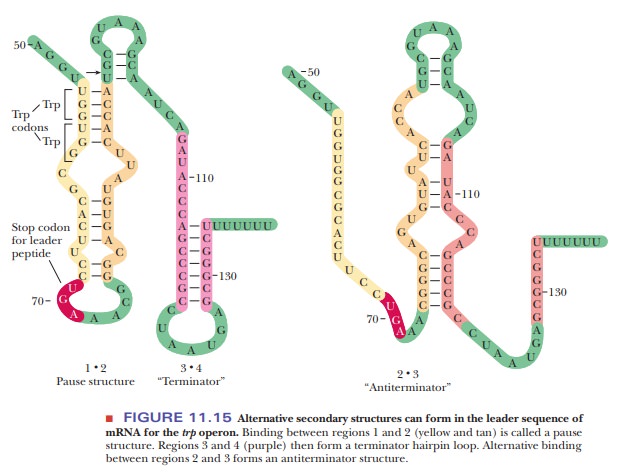
sequence are responsible for this effect (Figure 11.15).
How are RNA secondary structures involved in transcription attenuation?
Three
possible hairpin loops can form in this RNA-the 1?2 pause structure, the 3?4 terminator, or the 2?3 antiterminator.
Transcription begins normally and proceeds until position 92, at which point
the 1?2 pause structure can form. This causes RNA polymerase to pause in
its RNA synthesis. A ribosome begins to translate the leader sequence, which
releases the RNA polymerase from its pause and allows transcription to resume.
The ribosome follows closely behind the RNA polymerase shown in Figure 11.16.
The ribosome stops over the UGA stop codon of the mRNA, which prevents the 2?3
antiterminator hairpin from forming and allows instead the 3?4
terminator hairpin to form. This hairpin has the series of uracils
characteristic of rho-independent termination. The RNA polymerase ceases transcription
when this terminator structure forms.
If tryptophan is limiting, the ribosome stalls out over the tryptophan codons on the mRNA of the leader sequence.
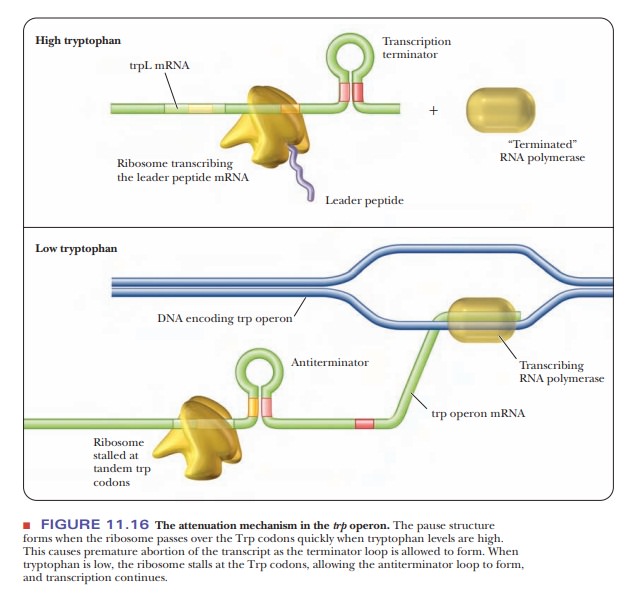
This leaves the mRNA free to form the 2’3
antiterminator hairpin, which stops the 3?4
terminator sequence from forming, so that the RNA polymerase continues to
transcribe the rest of the operon. Transcription is attenuated in several other
operons dealing with amino acid synthesis. In these cases, there are always
codons for the amino acid, which is the product of the pathway that acts in the
same way as the tryptophan codons in this example.
Summary
There are four principal control mechanisms for
prokaryotic transcription– alternative σ factors, enhancers, operons, and transcription
attenuation.
Alternative σ factors can direct RNA polymerase to different
promoters, altering the choice of RNA product.
Enhancers and silencers are
DNA sequences usually found upstream of promoters that stimulate or reduce
transcription, respectively. These sequences bind to specific proteins called
transcription factors.
Operons are groups of genes
involved in a common metabolic process that are controlled as a group. A common
example is the lac operon that produces
β-galactosidase
and other enzymes involved in metabolism of lactose.
With an operon, a regulatory
gene produces either an inducer or a repressor of the operon. A metabolite acts
as a co-inducer or co-repressor to affect the transcription of the structural
genes.
Transcription attenuation controls transcription after it has begun by adjust-ing the level of transcription based on the quantity of a related metabolite. For example, in the trp operon, the level of tryptophan affects the transcrip-tion of the genes that produce the enzymes that make tryptophan.
Related Topics