Chapter: Biochemistry: Transcription of the Genetic Code: The Biosynthesis of RNA
Transcription Regulation in Eukaryotes
Transcription Regulation in
Eukaryotes
In the
last section, we saw how the general transcription machinery, consisting of the
RNA polymerase and general transcription factors, functions to initiate
transcription. This is the general case that is consistent for all
transcription of mRNA. However, this machinery alone produces only a low level
of transcription called the basal level.
The actual transcription level of some genes may be many times the basal level.
The difference is gene-specific transcription factors, otherwise known as activators. Recall that eukaryotic DNA
is complexed to histone proteins in chromatin. The DNA is wound tightly around
the histone proteins, and many of the promoters and other regulatory DNA
sequences may be inaccessible much of the time.
Enhancers and Silencers
As seen
in prokaryotic transcription, enhancers and silencers are regulatory sequences
that augment or diminish transcription, respectively. They can be upstream or
downstream from the transcription initiator, and their orientation doesn’t
matter. They act through the intermediary of a gene-specific
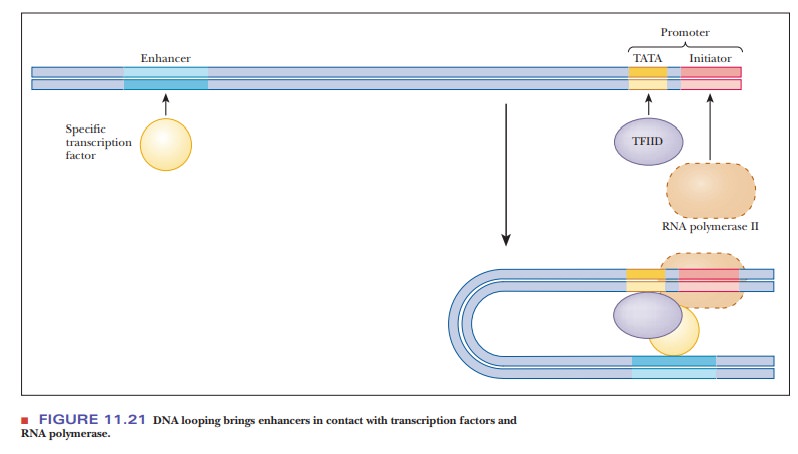
transcription-factor protein. As shown in Figure 11.21, the DNA must loop back
so that the enhancer element and its associated transcription factor can
contact the preinitiation complex. How this looping enhances transcription is
still unknown.
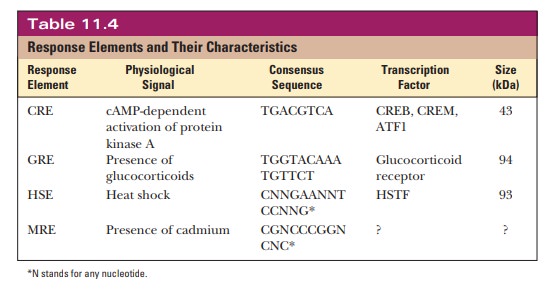
Response Elements
Some
transcription control mechanisms can be categorized based on a common response
to certain metabolic factors. Enhancers that respond to these factors are
called response elements. Examples
include the heat-shock element (HSE),
the glucocorticoid-response element
(GRE), the metal-response
element(MRE), and the
cyclic-AMP-response element (CRE).
How do response elements work?
These
response elements all bind proteins (transcription factors) that are produced
under certain cell conditions, and several related genes are activated. This is
not the same as an operon because the genes are not linked in sequence and are
not controlled by a single promoter. Several different genes, all with unique
promoters, may all be affected by the same transcription factor binding the
response element.
In the
case of HSE, elevated temperatures lead to the production of specific
heat-shock transcription factors that activate the associated genes.
Glucocorticoid hormones bind to a steroid receptor. Once bound, this becomes
the transcription factor that binds to the GRE. Table 11.4 summarizes some of
the best-understood response elements.
We will
look more closely at the cyclic-AMP-response element as an example of
eukaryotic control of transcription. Hundreds of research papers deal with this
topic as more and more genes are found to have this response element as part of
their control. Remember that cAMP was also involved in the control of
prokaryotic operons via the CAP protein.
Cyclic AMP is produced as a second messenger from several hormones, such as epinephrine and glucagon. When the levels of cAMP rise, the activity of cAMP-dependent protein kinase (protein kinase A) is stimulated. This enzyme phosphorylates many other proteins and enzymes inside the cell and is usually associated with switching the cell to a catabolic mode, in which macromolecules are broken down for energy.
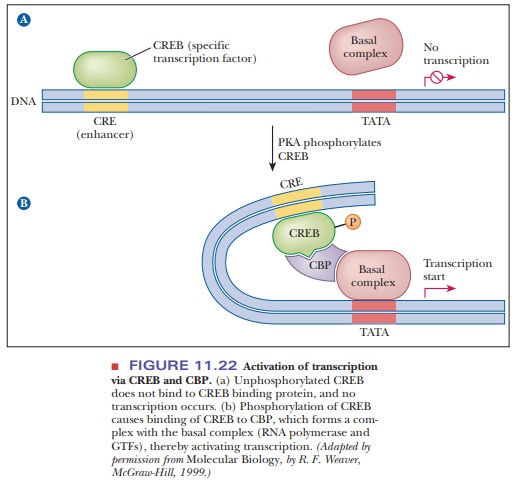
Protein kinase A phosphory-lates a
protein called cyclic-AMP-response-element
binding protein (CREB), which binds to the cyclic-AMP-response element and
activates the associated genes. The CREB does not directly contact the basal
transcription machinery (RNA polymerase and GTFs), however, and the activation
requires another protein. CREB-bindingprotein
(CBP) binds to CREB after it has been phosphorylated and bridges
theresponse element and the promoter region, as shown in Figure 11.22. After
this bridge is made, transcription is activated above basal levels. CBP is
called a mediator or coactivator. Many abbreviations are
used in the language of transcrip-tion, and Table 11.5 summarizes the more
important ones.
The CBP protein and a similar one called p300 are a major bridge to several dif-ferent hormone signals, as can be seen in Figure 11.23. Several hormones that act through cAMP cause the phosphorylation and binding of CREB to CPB. Steroid and thyroid hormones and some others act on receptors in the nucleus to bind to CBP/p300. Growth factors and stress signals cause mitogen-activated protein kinase(MAPK) to phosphorylate transcription factors AP-1 (activating protein 1) and Sap-1a, both of which bind to CBP.
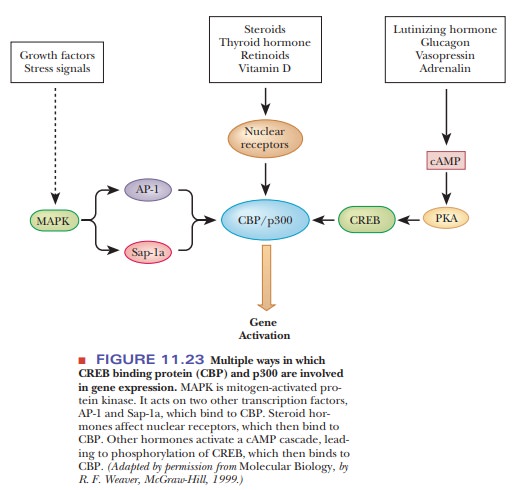
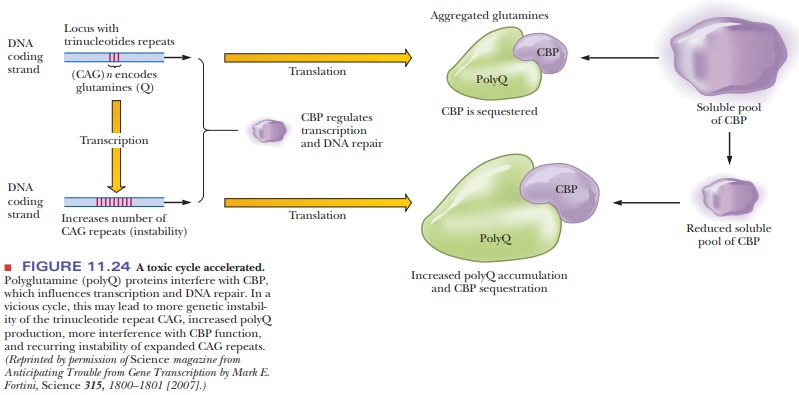
Researchers
have now linked several known human diseases to reduction of active levels of
CBP, including Huntington’s disease, spinocerebellar ataxia, and fragile X
syndrome, all of which are characterized by mutations that increase levels of a
trinucleotide repeat of CAG. This repeat produces polyglutamine when transcribed
and translated. The polyglutamine product sequesters CBP, making less of it
available for molecular processes, such as transcription and DNA repair. The
loss of the DNA repair leads to a propagation of the CAG repeats and leads to
the disease becoming worse in successive generations, a situation known as genetic anticipation. Figure 11.24
diagrams this syndrome.
RNA Interference
Although
it is convenient to think of transcription as being strictly relegated to one
strand of DNA, we are just now learning that there is a lot of transcription
from the opposite strand. RNA produced from the coding strand instead of the
template strand is noncoding RNA or antisense RNA. This type of RNA can produce
either micro RNA or small interfering RNA, and it can bind to the template
strand. This binding can either interfere with RNA polymerase binding or lead
to the outright destruction of targeted mRNA. Thus, the production of antisense
RNA is a new layer of gene regulation.
In late
2006, a California-based research team claimed to have found that small RNA
molecules can also be found to work as activators. Researchers were attempting
to use RNAi to block transcription of the human tumor suppres-sor gene,
E-cadherin. When they added synthetic RNAs designed to target the gene’s DNA,
they found instead that production of the tumor suppressor went up instead of
down. This process is tentatively being referred to as RNA activa-tion, or RNAa. It
is not clear at the moment whether this process was a positiveactivation of the
gene in question or an interference of some other gene that led to the indirect
activation of the E-cadherin gene. This process might be another powerful tool
in the scientist’s arsenal, as it would allow expanded ways to manipulate genes
and approach the fight against genetic diseases.
Summary
Control of eukaryotic transcription includes many of the same
concepts seen with prokaryotic transcription.
The use of enhancers and silencers is more extensive and the
promoters are more complicated.
Many control mechanisms are based on response
elements, enhancers that respond to some metabolic signal, such as heat, heavy
metals, or other specific molecules such as cAMP.
One very important response element is the
cyclic AMP response element (CRE). A specific transcription factor called CREB
and a mediating pro-tein called CBP are involved in many metabolic processes in
eukaryotes.
Small
noncoding RNAs make up a new layer of transcription control that is a hot topic
in molecular biology. These small RNAs bind to the tem-plate strand and inhibit
transcription.
Related Topics