Chapter: Genetics and Molecular Biology: Induction, Repression, and the araBAD Operon
Sugar Arabinose and Arabinose Metabolism
The Sugar Arabinose and Arabinose Metabolism
The
pentose L-arabinose occurs naturally in the walls of plant cells. The bacterium
E. coli, but not humans, can use this sugar as a source of carbon and
energy. Therefore arabinose is a free meal to intestinal flora when we eat a
meal containing vegetables. Before arabinose can be metabolized by the
bacterial intracellular arabinose enzymes, it must be transported from the
growth medium through the inner membrane to the cytoplasm. This is performed by
two independent arabinose transport systems, the products of the araE and araFGH genes. The araE
system possesses a low affinity for arabinose and therefore is most effective
in the presence of high concentrations of arabinose. The araFGH system possesses high affinity for arabinose uptake and
there-fore may be most valuable when arabinose concentrations are very low, on
the order of 10-7 M.
Arabinose,
like many sugars, exists in solution predominantly in a ring form in either of
two conformations. These interconvert with a half-time of about 10 minutes
(Fig. 12.1). Therefore it is possible that one of the transport systems is
specific for the α anomeric
form of arabinose and the other transports the β form. The β form is a substrate for the
first enzyme of the catabolic pathway, but it is not known whether the other
ring form or the linear form, which exists in only trace
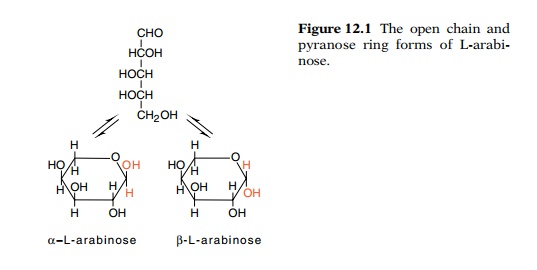
Figure
12.1 The open chain andpyranose ring
forms of L-arabi-nose.
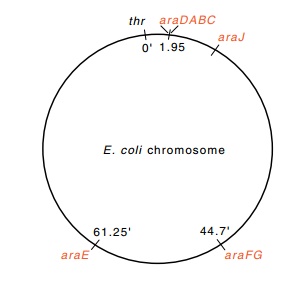
Figure
12.2 The locations on theE.
coli chromosome of the genesinduced by L-arabinose. Genes araE and araFGH are for uptake,
araBAD, for catabolism, and araJ isof
unknown function. The numbers indicate the gene positions on the genetic map.
The duplication of arabinose uptake systems hindered
their genetic and physiological study because mutants are then difficult to
identify. A defect in either system is masked by the activity of the other
system. The genes coding for the enzymes required for arabinose catabolism,
however, have been easier to map and study. They are located near the top of
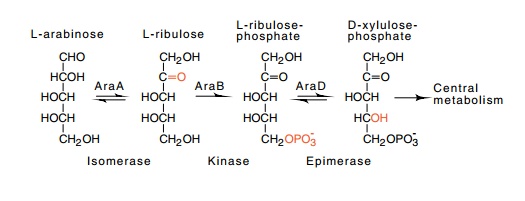
the genetic map as it is usually drawn (Fig. 12.2). Arabinose is first
converted to L-ribulose by arabinose isomerase, the araA gene product. Then ribulose is phosphorylated by ribulokinase,
the product of the araB gene, to
yield L-ribulose-5-phosphate (Fig. 12.3). The ribu-lose phosphate is next
converted to D-xylulose-5-phosphate by the araD
gene product, ribulose phosphate epimerase. Xylulose phosphate enters the
pentose phosphate shunt, and the enzymes subsequently involved are not induced
specifically by the presence of arabinose.
The genes of the two arabinose active transport
systems plus an additional arabinose-inducible gene of unknown function map to
three different regions of the chromosome. These are different from the map
locations of the genes required for the catabolism of arabinose. There-
Figure
12.3 The path of catabolism of
L-arabinose and the gene productscatalyzing the conversion.
fore a total of four sets of genes have been
discovered whose activities are regulated by arabinose. Why should this be? Why
not have all the genes in one large operon? Two possible reasons will be
discussed below.
At low arabinose concentrations, a high-affinity,
but perhaps energy-inefficient or low-capacity, uptake system might be
necessary for the cells to be able to utilize any arabinose at all. On the
other hand, in the presence of high concentrations of arabinose, a different
uptake system might be more useful. One with a high transport rate would be
neces-sary. This need not have a particularly high affinity for arabinose.
These different needs would necessitate splitting the corresponding genes into
separate operons so that they could be differentially regulated.
Alterna-tively, suppose arabinose were suddenly presented to bacteria, as it
might be in the gut. It is to the cell’s great advantage to begin metabo-lizing
this new nutrient just as soon as possible. If, however, all the genes for the
uptake and metabolism of arabinose were in a single long operon, then the
interval from induction until RNA polymerase could transcribe to the end of the
operon would be about three minutes. Any strain that divided its arabinose
operon into two or three separately transcribed units could induce all its ara enzymes more quickly and begin
apprecia-ble arabinose metabolism a minute or two sooner than cells with an
undivided arabinose operon. This time saved each time the operon was induced
could be of enormous selective value over evolutionary time scales.
Induction of the arabinose operon also requires the
presence of cyclic AMP and the cyclic AMP receptor protein CRP. The main role
of this protein is to enable induction of the arabinose operon only in the absence
of glucose. This prevents the cell’s attempting to utilize arabi - nose when
glucose, which is a better carbon source, is present. The general phenomenon of
being able to induce an operon well only in the absence of glucose is called
catabolite repression. A significant number of bacterial operons display
catabolite repression.
Related Topics